Methanococcus maripaludis: Difference between revisions
| Line 5: | Line 5: | ||
===Higher order taxa:=== | ===Higher order taxa:=== | ||
Archaea; Euryarchaeota; Methanococci; Methanococcales; Methanocaldococcaceae; Methanococcus | Archaea; Euryarchaeota; Methanococci; Methanococcales; Methanocaldococcaceae; Methanococcus | ||
===Species:=== | ===Species:=== | ||
Revision as of 18:54, 29 August 2007
Classification
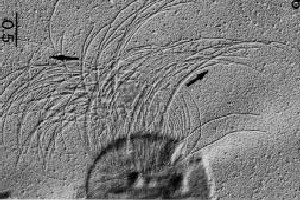
Higher order taxa:
Archaea; Euryarchaeota; Methanococci; Methanococcales; Methanocaldococcaceae; Methanococcus
Species:
Methanococcus Maripaludis
Description and significance
Methanococcus maripaludis is an irregular cocci shaped anaerobe that uses flagella for movement. It is mesophillic, meaning it grows under moderate conditions (20-45 degrees Celsius), and more specifically, its optimal growth temperature is 35-40 degrees Celsius. The species’ name maripaludis is latin for sea marsh, which is indicative of where the species can be found. This organism is aquatic and was originally isolated from a salt marsh sediment in South Carolina.
Methanococcus maripaludis is methanogenic, meaning it is a methane-producing archaeon. “Methanogens are obligate anaerobes that carry out the reduction of carbon dioxide to methane using molecular hydrogen as the reductant” (1). More specifically, Methanococcus maripaludis is categorized as hydrogenotrophic, using CO2 as its carbon source and H2 as a reducing agent. This means that this species undergoes anaerobic metabolic processes with the final product being methane, utilizing H2 as an electron donor for CO2 reduction to methane. Methanococcus maripaludis was the first genetically tractable hydrogenotrophic methanogenic species to be sequenced and can be used to genetically analyze the metabolism of other species in its class of methanogenic Archaea.
Methanococcus maripaludis is a significant microbe because it is an ideal model species because of fast reproducible growth and effective genetic tools. These methanogenic Archaea grow relatively rapidly under laboratory conditions and plate for single colonies at high efficiency. Genetic tools allow for microbiologists to analyze microbial DNA from microorganisms. Methanococcus maripaludis is helpful in “development of methods for growth on solid medium, enriching auxotrophic mutants, efficient transformation, and random insertional inactivation of genes” (2). It is because of these improvements to laboratory techniques that Methanococcus maripaludis is a popular archaebacteria for genetic manipulation.
Genome structure
The genome of Methanococcus maripaludis is similar to the genome of the more common Methanococcus jannaschii, with the difference being that Methanococcus maripaludis lacks inteins, DNA elements that are inserted in-frame and translated together with their host proteins. Methanococcus maripaludis contains 1,722 protein-coding genes in a single circular genome of 1.66Mb in length, with no extra-chromosomal elements. This chromosome also a 33% Guanine-Cytosine content (GC content) (3). In the genome, a nitrogen fixation (nif) gene cluster was characterized. Analysis of the sequence distinguished eight genes, six of which have similarity to known nif genes (4). This gene cluster is considered a “nitrogen regulon” and it contains genes for nitrogen metabolism that are regulated by a repressor binding site sequence, or operator (5). The genome for Methanococcus maripaludis is relatively condensed with only a few of its sequences are repeated.
Cell structure and metabolism
Methanococcus maripaludis is similar to Methanococcus voltae in its cell structure. “Methanococcus voltae is an archaebacterium and possess only a thin S-layer above the plasma membrane as its sole wall layer” (6). Also, similar to Methanococcus voltae, motility for Methanococcus maripaludis is via utilization of flagella, though the motility is weak. As previously stated, the shape of the cells of this species is irregular cocci.
Unlike other Methanococcus species, Methanococcus maripaludis is a nitrogen fixing species, meaning it takes in nitrogen from the environment and converts it into nitrogen compounds useful for chemical processes. Also, this species is able to use L- and D- Alanine as a nitrogen source utilizing alanine dehydrogenase and alanine racemase which are present in Archaea (7). Energy is generated via metabolic processes of this archaeabacteria. Methane, a flammable gas, is the final product of the anaerobic metabolic processes of Methanococcus maripaludis.
Ecology
“Methane formation occurs only under strictly anaerobic conditions” (8). Because of this, methanogenesis occurs only in environments that are anoxic, meaning environments that are abnormally low in or lacking oxygen. This species is strictly anaerobic, meaning it can live in the absence of oxygen. Organisms of the kingdom Archaea, in comparison to Eukaryotes, are able to grow under extreme conditions such as high temperature and high salt environments. Some Methanococcus species are commonly found in geothermal habitats such as thermal vents. As previously stated, Methanococcus maripaludis has been found in salt-marsh sediment on the southeastern coast of the United States.
Pathology
There is no known pathogen among different strains of Methanococcus maripaludis
Application to Biotechnology
As previously stated, methane is the final product of the anaerobic metabolic processes of Methanococcus maripaludis. This species is a commonly used archaeabacteria in laboratories for several reasons. A couple of reasons why it is a popular model is because of the fact that is capable of rapid growth and on plates it yields single colonies with high efficiency. Another reason why it is a model organism to study is because it has a relatively simple genome. Furthermore, this species has a large set of genetic tools. Integrative and expression vectors as well as positive and negative genetic selection are a few of the genetic tools for Methanococcus maripaludis.
Current Research
Currently, Methanococcus maripaludis is being studied for its mechanism of nitrogen regulation. Specifically, research is being done on the activity of nitrogenase, a nitrogen fixing enzyme in Methanococcus maripaludis. Also, research groups such as the John Leigh Lab of the University of Washington are studying hydrogen, the electron donor in the metabolic process involved in the production of methane, and its effects on regulating mRNA and protein levels in this species.
One field of research that Methanococcus maripaludis and other methanogens are involved in is astrobiology. Methanogenesis is currently being studied as a likely metabolism for any life that might exist on Mars. It has been reported that "certain methanogens can grow on a Mars soil simulant when supplied with carbon dioxide, molecular hydrogen, and varying amounts of water" (9). Because of this, methanogens can potentially exist below the surface of Mars, where a geothermal source of hydrogen, anaerobic water, and carbon dioxide is present.
Furthermore, since Methanococcus maripaludis already has an abundant set of genetic tools, genetic tool development is another aspect of this species that is being studied. Research groups are looking into other ways that this species can be helpful in genetic analysis and manipulation.
References
1. Dawes, Edwin. Microbial Energetics. New York: Blackie. 1986
2. Whitman et al. 1997. Development of genetic approaches for the methane-producing archaeabacterium Methanococcus maripaluis. Biofactors, Volume 6, Number 1: 37-46. http://www.ingentaconnect.com/content/ios/bio/1997/00000006/00000001/bio29;jsessionid=3ko9jad5p7srd.alice?format=print
3. Hendrickson, E., A. Haydock, B. Moore, W. Whitman, and J. Leigh. 2007. Functionally distinct genes regulated by hydrogen limitation and growth rate in methanogenic Archaea. Proc. Natl. Acad. Sci. USA 104:8930-8934.http://faculty.washington.edu/leighj/mm.html
4. Kessler PS, Blank C, Leigh JA. "The nif gene operon of the methanogenic archaeon Methanococcus maripaludis." Journal of Bacteriology. 1998. Volume 180. p 1504-11. http://www.ncbi.nlm.nih.gov/sites/entrez?Db=pubmed&Cmd=ShowDetailView&TermToSearch=9515920&ordinalpos=12&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum
5. Kessler PS, Leigh JA. "Genetics of nitrogen regulation in Methanococcus maripaludis." Genetics. 1999. Volume 152. p. 1343-51. http://www.ncbi.nlm.nih.gov/sites/entrez?Db=pubmed&Cmd=ShowDetailView&TermToSearch=10430565&ordinalpos=16&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum
6. Moat et al. Microbial Physiology, 4th edition. New York: Wiley-Liss. 2002
7. Hendrickson EL, Kaul R, Zhou Y, Bovee D, Chapman P, Chung J, Conway de Macario E, Dodsworth JA, Gillett W, Graham DE, Hackett M, Haydock AK, Kang A, Land ML, Levy R, Lie TJ, Major TA, Moore BC, Porat I, Palmeiri A, Rouse G, Saenphimmachak C, Söll D, Van Dien S, Wang T, Whitman WB, Xia Q, Zhang Y, Larimer FW, Olson MV, Leigh JA. "Complete genome sequence of the genetically tractable hydrogenotrophic methanogen Methanococcus maripaludis." Journal of Bacteriology. 2004. Volume 186. p.6956-69. http://www.ncbi.nlm.nih.gov/sites/entrez?cmd=Retrieve&db=pubmed&dopt=AbstractPlus&list_uids=15466049
8. Brock et al. Biology of Microorganisms, 7th edition. New Jersey. 1994
9. Timothy A. Kral, Curtis R. Bekkum and Christopher P. McKay. “Growth of Methanogens on a Mars Soil Simulant”. Origins of Life and Evolution of Biospheres. 2004. Volume 34. p. 615-626. http://www.springerlink.com/content/t5180771115805g4/
Edited by Allen De Leon, student of Rachel Larsen at UCSD.
KMG
