Methanococcus vannielii
Classification
Superkingdom: Archaea
Phylum: Euryarchaeota
Class: Methanococci
Order: Methanococcales
Family: Methanocaldococcaceae
Genus: Methanocaldococcus (syn: Methanococcus)
Species
|
NCBI: Taxonomy |
Genus species– Methanococcus vannielii
Description and Significance
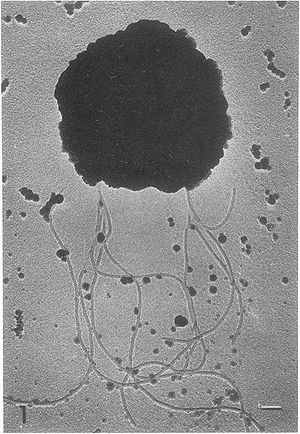
Methanococcus vannielii is a motile, coccus-shaped anaerobic microbe. Like some other Methanococcales, which are typically thermophiles, it is a mesophile, meaning it grows at an optimal temperature of 20 – 40 ºC (5). The name vannielii is Latin, meaning “from Niel”. This organism is aquatic and was originally isolated from a mud flat in San Francisco, California.
This microbe has significance in natural biogeochemical cycles of marine and wetland environments. Understanding these cycles and the role Archaea play can further our understanding of climate change. Methanogenesis also has implications as source of renewable bioenergy in engineered anaerobic digestion reactors. Further studies could indicate if Methanococcus vannielii is a competitive and efficient microbe for producing bioenergy.
Genome Structure
Funding for sequencing was provided by the US Department of Energy’s Joint Genome Institute (DOE JGI). Sequencing was completed in December, 2009. Methanococcus vannielii contains 1,701 protein-coding genes (less than M. maripaludis with 1,722 (2)). The total number of M. vannielii DNA bases is 1,720,048 (5). Only four out of the 13 rRNA genes contribute to 16S rRNA sequencing. One gene, responsible for the selenium binding protein, is unique to this organisms and has not been recorded in the gene bank. This protein is important for metabolism and growth by reducing harmful levels of selenium in the organism’s surroundings (6). Promotor regions in M. vannielii are found to contain TATA boxes homologous to those found in eukaryotes performing transcription(7). Methanococcus maripaludis was the first methanogenic species to be sequenced and can be used to compare and analyze the metabolism of other species of Archaea like M. vannielii.
Cell Structure, Metabolism and Life Cycle
Methanococcus vannielii is a free-living, gram-negative, coccus-shaped Archaea. It is nonsporulating and motile, using clusters of flagella, with cells arranged in a singular structure. The outer surface of the membrane is shown to contain several glycoprotein like structures similar to those found on halobacteria. These glycoproteins are typically used to maintain a definitive shape in the bacteria’s membrane. However, the membrane of M. vannielii is unable to maintain a rigid shape, does not require an osmotic gradient to maintain structure and ruptures readily upon centrifugation (4).
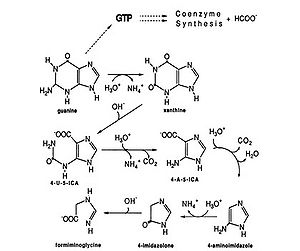
As an anaerobe, M. vannielii functions in anoxic environments. Methane-producing archaeon like Methanococcus vannielii are lithotrophic. “Methanogens are obligate anaerobes that can carry out the reduction of carbon dioxide to methane using molecular hydrogen as the reductant” (1). In addition to using formate as their main source of energy, M. vannielii metabolizes purines, especially guanines. In the presence of excess purines, M. vannielii degrades guanine to form formiminoglycine which can enter methanogenesis, by-passing the initial energy expensive step of reducing CO2 (3).
]
Ecology and Pathogenesis
M. vannielii inhabits anaerobic marine and mud environments. These environments have saturated soils with periodic flooding and varying depths. Organic matter accumulates from vegetation detris. In the San Francisco bay area marine mudflats have rapidly decreased in area due to urban and commercial development. Mudflat area in the bay reduced from 10% total aquatic area to 5.8% in 150 years (8).
The isolation project was located at Longitude -122.31075 and Latitude 37.69097 (5). Some Methanococcus species can be found in extreme habitats such as thermal vents, or very salty marsh sediment.
There are no known pathogens among Methanococcus vannielii.
]
References
(1) Dawes, Edwin. Microbial Energetics. New York: Blackie. 1986
(2) De Leon, Allen. 2007. Methanococcus maripaludis. Microbewiki. Kenyon University.
(3) DeMoil, E. "Purine metabolism in Methanococcus vannielii.." Journal of Bacteriology 175.18 (1993): 5754-5761. Web. 25 Apr 2010.
(4) Jones, Jay. "Methanococcus vannielii: Ultrastructure and Sensitivity to Detergents and Antibiotics." Journal of Bacteriology 130.3 (1977): 1357-1363. Web. 25 Apr 2010.
(5) Methanococcus vannielii. December 2009. Department of Energy Joint Genome Institute. The Regents of the University of California.
(6) Patteson, Kimberly. "Methanococcus vannielii selenium-binding protein (SeBP): Chemical reactivity of recombinant SeBP produced in Escherichia coli." Proceedings of the National Academy of the Sciences 102.34 (2005): 29-34. Web. 27 Apr 2010.
(7) Thomm, Michael. "An archaebacterial promoter element for stable RNA genes with homology to the TATA box of higher eukaryotes ." Oxford Journal 16.1 (1988): 151-163. Web. 27 Apr 2010.
(8) The Bay Institute. 1997. www.bay.org
Author
Page authored by Patrick Triscari and Brittany VanRaaphorst, students of Prof. Jay Lennon at Michigan State University.
<-- Do not remove this line-->