Micromonospora equina: Difference between revisions
No edit summary |
No edit summary |
||
| (27 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
{{Uncurated}} | {{Uncurated}} | ||
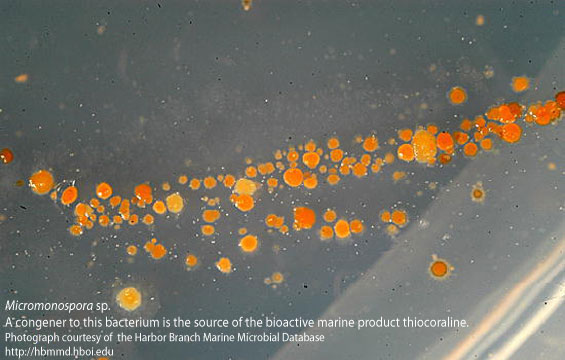
[[Image:Micromonospora_sp.jpg|frame|Microspora marina isolated from from coastal Mozambique, southeast Africa(http://www.marinebiotech.org/images_large/micromonospora_sp.jpg)]] | |||
==Classification== | ==Classification== | ||
| Line 14: | Line 16: | ||
==Description and Significance== | ==Description and Significance== | ||
The ''Micromonospora'' genus was discovered in 1923 by Øtdkov. The ''Micromonosporaceae'' family can be found in a variety of habitats including; freshwater or saltwater aquatic habitats, deep sea sediments, beach sand, root nodules, least common in soil. ''Micromonospora equina'' was discovered in the soil around a race horse track in Cape Town, South Africa. Two actinomycete strains (strain Y22T and strain Y21) were taken from this area. Strain Y21 colonies appear wrinkled and light orange in color. Strain Y22T colonies are a darker shade of orange. ''M. equina'' is a gram positive, spore forming, aerobic bacteria. Work is still being done to learn more about ''M. equnina'' but many strains of ''Micromonospora'' produce antibiotics such as gentamycin. While other strains can degrade rubber while others can survive water contaminated by radon, which is a radioactive by-product of uranium mining. | |||
==Genome Structure== | ==Genome Structure== | ||
Using rapid genus identification, researchers found that the isolates belong to one of six genera of ''Micromonosporaceae''. BLASTN analysis of 16srRNA gene on strain Y22 showed the microbe was in the genus ''Micromonospora'' and showed to have a very close sequence matches to ''Micromonospora viridifaciens'' DSM 43909T, ''Micromonospora auratinigra'' TT1-11T, and ''Micromonospora olivasterospora'' MK70T. | |||
==Cell Structure, Metabolism and Life Cycle== | ==Cell Structure, Metabolism and Life Cycle== | ||
This organism is a poor nitrate reducer that can produce H2S as a product. It is able to hydrolyze Aesculin, Arbitin and starch. The organism is able to degrade casein, gelatin, tween -80, and L-tyrosine. It is also able to inefficiently degrade xylan. The organism can use D-glucose, maltose, and threhalose as its only carbon source, and inefficiently us L-arabinose, α-lactose, and melibiose. ''M.equina'' can use L-asparagine, L-methionine, potassium nitrate, L-serine, L-histidine, L-4 hydroxyproline, and L-valine as its only nitrogen source. ''M. equina'' grows optimally at pH 5-10, 20-37ºC, but does not grow at 45°C. It can live at 2% NaCl, but one strain can live at 3% NaCl. The cell wall peptidoglycan is made up of DL- diaminopimelic acid (Meso-DAP) and glysine . Arabinose, glucose, ribose, and xylose was found in the cytoplasm, but there were no mycolic acids found. The polar lipids that were found in the membrane include phosphatidylethanilamine, phosphotidylinostitol, phosphatydylinostitol mannoside, and unidentified phospholipids. Strain Y22T was shown to be resistant to penicillin G. and weakly resistant to oleandomycin phosphate. This strain was sensitive to cephaloridine , lincomycin hydrochloride, neomycin sulfate, rifampicin, streptomycin sulfate, tobramycin sulfate, vancomycon hydrochloride. | |||
== | ==References== | ||
Ann M. Hirsch, Maria Valdés, Micromonospora: An important microbe for biomedicine and potentially for biocontrol and biofuels, Soil Biology and Biochemistry, Volume 42, Issue 4, April 2010, Pages 536-542, ISSN 0038-0717, 10.1016/j.soilbio.2009.11.023.(http://www.sciencedirect.com/science/article/pii/S0038071709004386) | |||
Gareth J., et al. 2013. Micromonospora equina sp.nov, isolated from soil from a racecourse. | |||
International Journal of Systematic and Evolutionary Microbiology 63:879-885. | |||
==Author== | ==Author== | ||
Latest revision as of 16:20, 20 April 2013
Classification
Bacteria; Actinobacteria; Actinobacteridae; Actinomycetales; Micromonosporineae; Micromonosporaceae; Micromonospora NCBI link to find]
Species
|
NCBI: Taxonomy |
Micromonospora equina
Description and Significance
The Micromonospora genus was discovered in 1923 by Øtdkov. The Micromonosporaceae family can be found in a variety of habitats including; freshwater or saltwater aquatic habitats, deep sea sediments, beach sand, root nodules, least common in soil. Micromonospora equina was discovered in the soil around a race horse track in Cape Town, South Africa. Two actinomycete strains (strain Y22T and strain Y21) were taken from this area. Strain Y21 colonies appear wrinkled and light orange in color. Strain Y22T colonies are a darker shade of orange. M. equina is a gram positive, spore forming, aerobic bacteria. Work is still being done to learn more about M. equnina but many strains of Micromonospora produce antibiotics such as gentamycin. While other strains can degrade rubber while others can survive water contaminated by radon, which is a radioactive by-product of uranium mining.
Genome Structure
Using rapid genus identification, researchers found that the isolates belong to one of six genera of Micromonosporaceae. BLASTN analysis of 16srRNA gene on strain Y22 showed the microbe was in the genus Micromonospora and showed to have a very close sequence matches to Micromonospora viridifaciens DSM 43909T, Micromonospora auratinigra TT1-11T, and Micromonospora olivasterospora MK70T.
Cell Structure, Metabolism and Life Cycle
This organism is a poor nitrate reducer that can produce H2S as a product. It is able to hydrolyze Aesculin, Arbitin and starch. The organism is able to degrade casein, gelatin, tween -80, and L-tyrosine. It is also able to inefficiently degrade xylan. The organism can use D-glucose, maltose, and threhalose as its only carbon source, and inefficiently us L-arabinose, α-lactose, and melibiose. M.equina can use L-asparagine, L-methionine, potassium nitrate, L-serine, L-histidine, L-4 hydroxyproline, and L-valine as its only nitrogen source. M. equina grows optimally at pH 5-10, 20-37ºC, but does not grow at 45°C. It can live at 2% NaCl, but one strain can live at 3% NaCl. The cell wall peptidoglycan is made up of DL- diaminopimelic acid (Meso-DAP) and glysine . Arabinose, glucose, ribose, and xylose was found in the cytoplasm, but there were no mycolic acids found. The polar lipids that were found in the membrane include phosphatidylethanilamine, phosphotidylinostitol, phosphatydylinostitol mannoside, and unidentified phospholipids. Strain Y22T was shown to be resistant to penicillin G. and weakly resistant to oleandomycin phosphate. This strain was sensitive to cephaloridine , lincomycin hydrochloride, neomycin sulfate, rifampicin, streptomycin sulfate, tobramycin sulfate, vancomycon hydrochloride.
References
Ann M. Hirsch, Maria Valdés, Micromonospora: An important microbe for biomedicine and potentially for biocontrol and biofuels, Soil Biology and Biochemistry, Volume 42, Issue 4, April 2010, Pages 536-542, ISSN 0038-0717, 10.1016/j.soilbio.2009.11.023.(http://www.sciencedirect.com/science/article/pii/S0038071709004386)
Gareth J., et al. 2013. Micromonospora equina sp.nov, isolated from soil from a racecourse. International Journal of Systematic and Evolutionary Microbiology 63:879-885.
Author
Page authored by Chanel Redden and Stephanie DeDore, students of Prof. Edward Walker at Michigan State University.
<-- Do not remove this line-->
