Mycobacterium smegmatis: Difference between revisions
| Line 25: | Line 25: | ||
==Genome structure== | ==Genome structure== | ||
[[Image:Ncgraph.png|thumb|500px|left| A cartoon of the ''Mycobacterium smegmatis'' gene. | [[Image:Ncgraph.png|thumb|500px|left| A cartoon of the ''Mycobacterium smegmatis'' gene. From [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=genome&cmd=Retrieve&dopt=Overview&list_uids=20087 NCBI Entrez Genome Website.]]] | ||
The genome is 6,988,209 nucleotides long. It has a 67% GC content and a 33% AT content, and is therefore classified as a high GC content gram positive bacteria (discussed below). 90% of the genome (6716/6938 genes) represents coding regions that encode for 6716 proteins. The 6938 genes that are composed circularly with an absence of any plasmids. ''Mycobacterium smegmatis'' is a slow growing bacteria which contains one copy of the ribosomal RNA genes unlike fast growing bacteria (Eg. ''Escherichia coli'') which has two copies of the rRNA genes. ''Mycobacterium smegmatis'' doesn't need so many copies of the genes because it doesn't require the high production of proteins when it is growing slow, while ''Escherichia coli'' does. The genome was sequenced in November 29, 2006 by the J. Craig Venter Institute. 9. | The genome is 6,988,209 nucleotides long. It has a 67% GC content and a 33% AT content, and is therefore classified as a high GC content gram positive bacteria (discussed below). 90% of the genome (6716/6938 genes) represents coding regions that encode for 6716 proteins. The 6938 genes that are composed circularly with an absence of any plasmids. ''Mycobacterium smegmatis'' is a slow growing bacteria which contains one copy of the ribosomal RNA genes unlike fast growing bacteria (Eg. ''Escherichia coli'') which has two copies of the rRNA genes. ''Mycobacterium smegmatis'' doesn't need so many copies of the genes because it doesn't require the high production of proteins when it is growing slow, while ''Escherichia coli'' does. The genome was sequenced in November 29, 2006 by the J. Craig Venter Institute. 9. | ||
Revision as of 02:44, 4 June 2007
A Microbial Biorealm page on the genus Mycobacterium smegmatis
Classification
Higher order taxa
Bacteria (Domain); Actinobacteria (Phylum); Actinobacteridae (Class); Actinomycetales (Order); Corynebacterineae (Suborder); Mycobacteriaceae(Family); Mycobacterium (Genus). 8.
Species
Mycobacterium smegmatis.
Also known by: Mycobacterium paratuberculosis smegmatis, Bacterium smegmatis, Bacillus smegmatis, Mycobacterium paratuberculosis smegmatis, Bacterium smegmatis, Bacillus smegmatis, Mycobacterium smegmatis. 7.
Description and significance
Mycobacterium smegmatis was first discovered in 1884 by Lustgarten. They are mostly found in the soil, marine, and freshwater environments. Mycobacterium smegmatis is classified as a saprophytic species that rarely causes disease and isn't dependent on living in an animal, unlike some pathogenic Mycobacterium.
Mycobacterium smegmatis is a very useful for the research analysis of other species in the genus Mycobacteria in cell culture laboratories. There are several Mycobacterium species that are common, harmful diseases, like Mycobacterium leprea, Mycobacterium tuberculosis, and Mycobacterium bovis. Mycobacterium smegmatis is so important because it is fast growing and non-pathogenic compared to these species. There are many similarities between Mycobacterium smegmatis and the much more virulent obligate pathogens that are Mycobacteria. The most significant is the complementary uses of mycothiol biosynthesis of Mycobacterium for making an essential thiol that is responsible for life. If it is knocked out, the species will be terminated and a treatment will be found. 10. There is also research involved in finding drug therapies that will treat inhibit the myolic acid biosynthesis which is essential for creating the unique bacterial cell wall. 11. Currently, there are many laboratories that are culturing and isolating this species to determine the pathological course of deleterious Mycobacteria.
Describe the appearance, habitat, etc. of the organism, Describe how and where it was isolated. Include a picture or two (with sources) if you can find them.
Genome structure
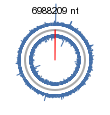
The genome is 6,988,209 nucleotides long. It has a 67% GC content and a 33% AT content, and is therefore classified as a high GC content gram positive bacteria (discussed below). 90% of the genome (6716/6938 genes) represents coding regions that encode for 6716 proteins. The 6938 genes that are composed circularly with an absence of any plasmids. Mycobacterium smegmatis is a slow growing bacteria which contains one copy of the ribosomal RNA genes unlike fast growing bacteria (Eg. Escherichia coli) which has two copies of the rRNA genes. Mycobacterium smegmatis doesn't need so many copies of the genes because it doesn't require the high production of proteins when it is growing slow, while Escherichia coli does. The genome was sequenced in November 29, 2006 by the J. Craig Venter Institute. 9.
Cell structure and metabolism
Mycobacterium smegmatis is a Gram-positive bacteria, characterized by an inner cell membrane and a thick cell wall. The Gram-positive bacteria is further classified as one with a high GC content and therefore a low AT content. This quality is used as a rude measure of similarity of different species of bacteria. Although this bacteria is Gram-positive, its cell wall contains mycolic acids, long, branched fatty acids that are normally present in Acid-fast bacteria. The acids prevent proper gram staining that would normally identify the cell as a gram positive cell because they create a waxy coating so the crystal violet has difficulty entering the cell, therefore making it seem gram negative. The cell wall is also abnormal because it is irregularly thick for a gram positive bacteria and its hydrophobicity reduces dessication. This feature in addition to its slow cell growth attribute to Mycobacterium smegmatis' low response to antibiotics.
Although it contains the similar structural features of Mycobacterium Tuberculosis, it grows much quicker in comparison. Mycobacterium smegmatis is an aerobic organism. Small amounts of Mycobacterium smegmatis may also survive on chemolithoautotrophic growth. They would use carbon dioxide and hydrogen gas as its inorganic fuel source. It also requires a unique fatty acid biosynthesis to produce the mycolic acids that are present on the cell wall.
Ecology
Mycobacterium smegmatis lives in aggregate layers of cells attached to each other in a community called a biofilm. may live in soil and degrade organic materials, including sterols. The bacteria lives in water, and tends to exist near large bodies of water.
Describe any interactions with other organisms (included eukaryotes), contributions to the environment, effect on environment, etc.
Pathology
This organism is classified as saprophytic and therefore extremely unpathogenic. Mycobacterium smegmatis doesn't normally reside in any animals, and doesn't cause dangerous or even any infections. There are no threats as it is very safe. However other species under this genus are pathogenic. Obligate pathogens such as Mycobacterium tuberculosis, Mycobacterium bovis, and Mycobacterium leprea are highly pathogenic in animals. These species can cause tuberculosis and leprosy. There is also the classification of potential pathogens in this genus, such as Mycobacterium avium.
However, there has been 1 case where the bacteria has killed a human after infection. In this case, an Italian child was infected by Mycobacterium smegmatis and died at the age of 8. After genetic analysis, it was determined that four nucleotides were inserted, causing a frameshift and a nonsense mutation. The stop codon allowed for a premature stop of translation. Although this showed potential pathogenic properties of Mycobacterium smegmatis, this case required that the child have two mutant alleles in his genome for the bacteria to be particularly virulent. Therefore in most cases, it is generally safe. 5.
Application to Biotechnology
Mycobacterium smegmatis has been used to produce Xylitol. Xylitol is an important sugar that is used as a substitute for commercial dietary sugars to prevent tooth decay. It is also a plaque reducer. It is also absorbed more slowly in the bloodstream, so it prevents the negative affects of high blood sugar levels. This is used as a dietary alternative for diabetics. It also has potential treatments for osteoporosis and ear and upper respiratory infections. The D-xylulose sugar produced by Mycobacterium smegmatis is combined with the D-xylose sugar produced by D-xylose isomerase of immobilized Mycobacterium smegmatis to produce the beneficial Xylitol. 6.
Mycobacterium smegmatis is also used to transform L-ribulose into L-arabinose with the enzyme L-arabinose isomerase. Mycobacterium smegmatis' produces L-arabinose during carbohydrate metabolism. L-arabinose is used by biotech companies to produce chiral drugs. L-arabinose is also used as a component for many tissue culture medium. It is also used by food producers for the Maillard reaction to make bread, beer, dried or condensed milk, etc. 6.
Current Research
Dr. Fahey's lab at UCSD studies the methods of mycothiol biosynthesis that produces the thiols that are needed for Mycobacterium to live and grow. The lab is trying to determine the missing and unknown enzymes and substrates that are present in the biosynthesis and if they are essential. There are currently three steps that have been determined by this lab. They are trying to determine the unknown substrate that is converted to GlcNAc-Ins from the enzyme MshA. From 3 more known steps using MshB, MshC, and MshD, the necessary mycothiol is produced. The Fahey lab aims to determine the unknown substrate to determine the full mycothiol biosynthesis pathway, and to determine some inhibitors of this pathway to prevent Mycobacterium smegmatis growth, which can also translate to treatments for Mycobacterium tuberculosis. 2
Mycobacterium smegmatis uses a terminal oxidase to donate electrons to the final electron acceptor oxygen in oxidative phosphorylation during aerobic respiration. These terminal oxidases have been identified as using both a cytochrome c aa3 type oxidase and a quinol bd type oxidase. When bd type oxidase gene is knocked out to only allow for the cytochrome c aa3 type oxidase to function, the latter didn't attain normal levels of expression of the oxidase. In addition, the oxidase concentration for the knockout didn't reduce during log phase, while the wild type(without the knockout) had decreased oxidase concentration. 3
Mycobacteria smegmatis is also dependent on the import of free inorganic phosphate molecules. Genes that regulate this import are often two component regulatory systems that are composed of histidine kinase and a DNA-binding response regulator. One of these regulators discovered by Dr. Glover is senX3-regX3 2CR. SenX3 acts as a phosphotase and a phosphate donor for regX3. A phosphorylated regX3 will activate phosphate acquisition. RegX3-P is bound to the promoter regions and activates the transcription of senX3, phoA, pstS genes that activate inorganic phosphate import. Therefore this group has determined that the senX3-regX3 2CR system is responsible for regulating this pathway. 4
References
1. Dastur, A., Kumar, P., Ramesh, S., Vasanthakrishna, M., Varshney, U. "Analysis of the initiator tRNA genes from a slow- and a fast-growing Mycobacterium" Department of Microbiology and Cell Biology, Indian Institute of Science, Bangalore, 560 012, India. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=12209262&dopt=Abstract
2. Newton, G., Ta, P., Bzymek, K., Fahey, R. "Biochemisrty of the Initial Steps of Mycothiol Biosynthesis" Journal of Biological Chemistry. 2006. Volume 281. p. 33910-20. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=16940050&query_hl=9&itool=pubmed_docsum
3. Megehee, J., Lundrigan, M. "Temporal expression of Mycobacterium smegmatis respiratory terminal oxidases" Canadian Journal of Microbiology. 2007. Volume 53. p. 459-6. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=17538658&query_hl=5&itool=pubmed_docsum
4. Glover, R., Kriakov, J., Garforth, S., Baughn, A., Jacobs, W. "The Two-Component Regulatory System senX3-regX3 Regulates Phosphate-Dependent Gene Expression in Mycobacterium smegmatis." Journal of Bacteriology. 2007. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=17526710&query_hl=5&itool=pubmed_docsum
5. Branca, A., Baglioni, C. "Evidence that types I and II interferons have different receptors." Nature. 1981. Volume 294: p. 768-770. http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=107470
6. Ahmed, Z. "Production of natural and rare pentoses using microorganisms and their enzymes." Electronic Journal of Biotechnology. 2001. Volume 4. Number 2.
9. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=genome&cmd=Retrieve&dopt=Overview&list_uids=20087
10. Newton, G., Fahey, R. "Mycothiol biochemistry." Arch Microbiol. 2002. Volume 178. p. 388-94. http://www.springerlink.com/content/0jvtmpat4863q0vk/
11. Schroeder, E., de Souza, N., Santos, D., Blanchard, J., Basso, L. "Drugs that inhibit mycolic acid biosynthesis in Mycobacterium tuberculosis." Curr Pharm Biotechnol. 2002. Volume 3. p. 197-225. http://www.ingentaconnect.com/content/ben/cpb/2002/00000003/00000003/art00002?token=004816d0ad50b6726e2d2954496f642f466f256720297d7625627b507b5f5f316a6f3874
Edited by Benjamin Yip, student of Rachel Larsen and Kit Pogliano
