Natranaerobius thermophilus
Classification
Domain: Bacteria
Phylum: Firmicutes
Class: Clostridia
Order: Natranaerobiales
Family: Natranaerobiaceae
Species
Natranaerobius thermophilus
NCBI Taxonomy ID: 59001
Description and Significance
First discovered and isolated in 2005, N. thermophilus was characterized as the first identified truly anaerobic, halophilic alkalithermophilic organism. The bacterium is a rod-shaped, non-motile, non-sporeforming bacteria. It is an obligate anaerobe and Gram-positive.
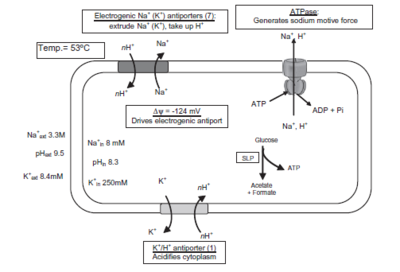
Being a polyextremophile, it thrives in multiple extremes of the environment. The bacteria are obligate alkaphiles and obligate halophiles. As such, N. thermophilus has various growth optima. Its preferred temperature is 53°C, with a culturable temperature range of 30-57°C. Its pH optima is 9.5, with a range of pH 8.5-10.6, at 55°C. Optimal growth occurs with a salt concentration of around 3.3-3.9 M Na+ (range 1.5-4.9 M Na+). This polyextremophile provides significant scientific insight as to the strategies it uses for sirvival.
Genome Structure
The genome of N. thermophilus was sequenced in 2011 and consists of one circular 3.16 mbp chromosome and two smaller plasmids, one being 17.2 kbp and the other 8.69 kbp. The G+C content of the whole genome was found to be 36.4%. Its plasmids, pNTHE01 and pNTHE02 have GC contents of 34.1% and 35.7%, respectively. [4]
Many genes were recognized that are believed to be associated with the bacteria's polyextremophile capabilities. From the genome sequence analysis, there are many proteins that allow for life under such extreme conditions: alkaline pH, elevated temperature, and high salinity. Three genes for L-glutamine synthetase, 15 genes for glycine betaline ABC transporters, four genes for glycine betaine/L-proline ABC transporters, and three genes for betaine/carnitine/chlorine transporters have been found, as well as the gsmA gene and the sdmA gene that endode for de novo synthesis of solute glycine betaine. [4]
Cell Structure, Metabolism and Life Cycle
The cell structure are straight to curved rods, 0.2-0.4 µm in diameter, and 3-5 µm in length. Cells either aggregated into chains or were found singular and non-motile.
Some of the carbon and energy sources utilized by N. thermophilus are: tryptone, fructose, ribose, pyruvate, trimethylamine, acetate, xylose, peptone, among others. Its main organic fermentation products from sucrose are formate and acetate. It utilizes fumarate, sulface, nitrate, and iron(III) citrate as electron acceptors. [3]
Many of the extremophile capabilities of N. thermophilus are attributed to the membrane transport proteins, like anitporters and symporters. "The genome sequence analysis revealed five genes that encode Na+/proline symporters, two for Na+/glutamate symporters, and seven for K+ transport systems, which together with a specific K+/H+ antiporter regulate the intracellular K+ concentration." [4] There were also seven sodium and potassium proton antiporters found, with a K+ antiporter and 3 K+ transporters being experimentally verified.
The extreme halophilic alkaline conditions that N. thermohiilus lives in require that it has mechanisms for cytoplasm acidification. To achieve this, its chromosome "contains genes for 11 monovalent cation/proton antiporters of the NhaC type, a gene cluster encoding a multisubunit cation/proton antiporter of the CPA-3 family, four monovalent cation/proton antiporters of the CPA1 and CPA2 family, and one gene encoding a cation/proton antiporter of the NdhF-a family." [4]
In order to maintain structural stability, N. thermophilus was found to contain four genes that encoded orthologous rRNA MTases and three orthologous tRNA MTases. These assist with stabilization of DNA and RNA at high temperatures. 12 heat shock proteins were also identified.
Ecology and Pathogenesis
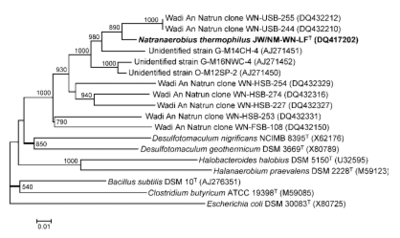
Original discovery of N. thermophilus was isolated from the sediment in an alkaline, hypersaline, Lake Fazda in Wadi An Natrun, Egypt, in 2005. The salinity of the water in the lake was 4.7 M and had a pH of 9.8. At the time, the family, order, genus, and species identified were all novel lineages discovered.
As this organism utilizes unique combinations of adaptation mechanisms, it is considered to be an excellent model organism. From this bacterium, scientists have been able to further study anaerobic halophilic alkalithermophiles and their features that allow them to thrive in a vast array of environmental extremes.
No known pathogenicity has been determined in this organism.
References
Author
Page authored by Zach Geurin, Caroline Moon, and Mike Reitmeyer, student of Prof. Jay Lennon at Michigan State University.
<-- Do not remove this line-->