Pediococcus damnosus: Difference between revisions
(Created page with "==Classification== ==Description and Morphology== ======Pedicocci are found in fruit, fermented foods and plant material. Pediococcus damnosus is gram-positive bacteria that...") |
No edit summary |
||
| (48 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
{{Uncurated}} | |||
==Classification== | ==Classification== | ||
== | ===Higher order taxa=== | ||
======Pedicocci are found in fruit, fermented foods and plant material. Pediococcus damnosus | |||
Bacteria; Bacteria; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; ''Pediococcus'' | |||
[[image:Pediococcus.gif]] | |||
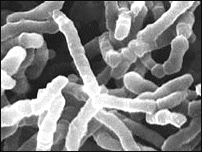
'''Figure 1-1: Morphology of ''Pediococcus damnosus''''' | |||
===Species=== | |||
''Pediococcus damnosus'' | |||
==Description and Significance== | |||
''Pedicocci'' are found in fruit, fermented foods and plant material. ''Pediococcus damnosus'' are gram-positive bacteria that are obligate anaerobes metabolizing sugars among carbohydrates resulting in fermentation. Its morphology displays short chains of cocci cells (0.3-0.6 mm in diameter). Round colonies express an off white/cream pigmentation while spores, capsules and motility are all absent. '''Figure 1-1''' is a malolactic fermentation bacterium that shows ''Pediococcus'' morphology. | |||
==Growth== | |||
Besides food and plant material, glucose, sucrose and galactose are metabolized by this bacterium. Some strains will ferment maltose and sucrose as well. This particular species is facultative allowing the metabolic processes to undergo with or without oxygen. With glucose, sucrose and galactose being primary targets for this microbe, it is one of the most common and well-known spoilers of wines, ciders, beers and juices. Other common microbes responsible for beer and wine spoilage include Acetobacter, Gluconobacter, Megasphaera cerevisiae, Saccharomyces diastaticus and Candida valida. Faster detection of ''Pediococcus damnosus'' and other contaminating microbes will help prevent the spoilage of produced beverages and stop the unnecessary loss of countless dollars for their companies. (Brodmann, 2011) | |||
==Lactic Acid Bacteria== | |||
Bacteria found among food spoilage and food-borne pathogens are abundant. Lactic acid bacteria (LAB) are found among ''Pediococcus damnosus''. Experiments have proven that this bacterium is associated with both food and beverage spoilage. More so, Lactobacillus phage B1 genome has 97 percent identity with ''Pediococcus damnosus''; ensuring that it affects spoilage just as lactic acid bacteria does (Marco, 2012). | |||
=== Lactic Acid Bacteria In Wine === | |||
Lactic acid bacteria play an important role for wine quality. A species-specific multiplex PCR system was performed to detect the presence of lactic acid bacteria species in ''Pediococcus damnosus''. DNA extraction, multiplex PCR and the analysis of results prove that LAB is present in this microbe. The strain shows extreme resistance to ethanol and other influences so they are likely to appear during or after malolactic fermentation (Petri, 2012). | |||
Companies have benefited from researchers developing a rapid method that can be used for detection, quantification, and identification of this microorganism. GC-MS and other slow cultivation and microscopy techniques are being replaced by analytical methods and test kits that that only take 12 hours to complete (Brodmann, 2011). These new methods look for certain DNA sequences in order to determine the presence of contamination, and if so, how much and which species. | |||
[[image:Pediococcus2.jpeg]] | |||
'''Figure 2-1: Wine fermentation''' | |||
Some strains of microorganisms are capable of surviving and multiplying rapidly even after the fermentation and sulfating processes of wine production. There are strict microbiological regulations and qualifications that beverages must pass before being sold so contamination is a costly problem. '''Figure 2-1''' shows the start of fermentation in different flavors of wine. Certain strains of ''Pediococcus damnosus'' can synthesize β-D-glucan, a glucose homopolymer that changes the viscosity of wine to be thicker than oil (Delaherche, 2004). These microorganisms used to be detected and quantified by selective counting on MRS plates, where the colony-growth step took seven days. Researchers have now developed a more time-efficient method (taking one day) using real-time quantitative PCR as a new technique that can be used for routine screening (Delaherche, 2004). | |||
==Laboratory Studies== | |||
''Pediococcus damnosus'' produces exopolysaccharides that change the viscosity of and ruin products. Primers PF5 and PF6 have been analyzed via PCR, along with new primers PF1 and PF8 that target the dps gene (putatative glucan synthase gene). This gene was found vastly among isolated strains of both ''Pediococcus damnosus'' and ''Oenococcus oeni'' (Walling, 2005). In order to ascertain that the cells were indeed producing the wine-spoiling glucan, an aggregation test (binding test) was performed with ''Streptococcus pneumoniae'' type 37-specific antibodies. Agglutination proved that there is a direct relation between gene dps and glucan production (Walling, 2005). This shows that gene dps could be the cause of exopolysaccharide production not only in ''Pediococcus damnosus'', but in ''Oenococcus oeni'' as well. | |||
The microbe is further studied with different controls to determine resistance and sensitivities. Pediocin PD-1 is one, of many studied, bactericidal antimicrobial peptides. ''Pediococcus damnosus'' produces pediocin PD-1 and so researchers find this microbe to be timely. A gel electrophoresis experiment was used to detect pediocin PD-1 to be a molecular mass of 35 kDa. Researchers continued to undergo experiments with pediocin PD-1 determining the isoelectric point and antimicrobial activity. Results from their tests revealed that pediocin PD-1 is also resistant to specific treatments (Green, 1997). | |||
While most microbes are harmless to humans ''Pedicococcus damnosus'' is yet undesired, specifically for beer and wine companies. It is considered a spoilage bacterium in wine making and brewing fields that also contributes to unwanted viscosity due to glucan synthesis. Researchers have supplied fast detection, quantification, and identification of this microbe to help the major distributing company’s preserve their products. | |||
==References== | |||
[http://www.google.com/patents/CA2571646A1]Brodmann, Peter D, Fehlmann M, Gafner J, Methods and Kits Pertaining to the Detection, Identification and Quantification of Bacteria and Yeast in Wine, Beer, and Juices. ETS Laboratories, assignee. Patent US 8,017,359 B2. 13 Sept. 2011. | |||
[http://onlinelibrary.wiley.com/doi/10.1111/j.1365-2672.2004.02334.x/pdf]Delaherche A, Claisse O, Lonvaud-Funel A, "Detection and Quantification of ''Brettanomyces bruxellensis'' and 'ropy' ''Pediococcus damnosus'' Strains in Wine by Real-time Polymerase Chain Reaction." Journal of Applied Microbiology 97.5 (2004): 910-15. | |||
[http://onlinelibrary.wiley.com/doi/10.1046/j.1365-2672.1997.00241.x/pdf]Green G, Dicks L.M.T, Bruggeman G, Vanamme E.J, Chikindas M.L 1997. Pediocin PD-1, a bactericidal antimicrobial peptide from ''Pediococcus damnosus'' NCFB 1832. Journal of Applied Microbiology(83): 127-132. | |||
[http://aem.asm.org/content/78/24/8719.full.pdf+html]Marco M, Garneau J, Tremblay D, Quiberoni A, Moineau S 2012. Characterization of Two Virulent Phages of ''Lactobacillus plantarum''. Applied and Environmental Microbiology (78):8719-8734. | |||
[http://ac.els-cdn.com/S0740002012001761/1-s2.0-S0740002012001761-main.pdf?_tid=8cfef2d2-6361-11e3-89d0-00000aacb361&acdnat=1386875818_17a383b6e44ec086fbb66e40b57e0685]Petri A, Pfannebecker J, Frohlich J, Konig H 2012. Fast identification of wine related lactic acid bacteria by multiplex PCR. Food Microbiology (33): 48-54. | |||
[http://www.sciencedirect.com/science/article/pii/S016816050400279X]Walling E,Gindreau E, Lounvaud-Funel A, "A Putative Glucan Synthase Gene dps Detected in Exopolysaccaride-Producing ''Pediococcus damnosus'' and ''Oenococcus oeni'' Strains Isolated From Wine and Cider." International Journal of Food Microbiology (2005): 53-62. | |||
Latest revision as of 21:51, 1 December 2015
Classification
Higher order taxa
Bacteria; Bacteria; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Pediococcus
Figure 1-1: Morphology of Pediococcus damnosus
Species
Pediococcus damnosus
Description and Significance
Pedicocci are found in fruit, fermented foods and plant material. Pediococcus damnosus are gram-positive bacteria that are obligate anaerobes metabolizing sugars among carbohydrates resulting in fermentation. Its morphology displays short chains of cocci cells (0.3-0.6 mm in diameter). Round colonies express an off white/cream pigmentation while spores, capsules and motility are all absent. Figure 1-1 is a malolactic fermentation bacterium that shows Pediococcus morphology.
Growth
Besides food and plant material, glucose, sucrose and galactose are metabolized by this bacterium. Some strains will ferment maltose and sucrose as well. This particular species is facultative allowing the metabolic processes to undergo with or without oxygen. With glucose, sucrose and galactose being primary targets for this microbe, it is one of the most common and well-known spoilers of wines, ciders, beers and juices. Other common microbes responsible for beer and wine spoilage include Acetobacter, Gluconobacter, Megasphaera cerevisiae, Saccharomyces diastaticus and Candida valida. Faster detection of Pediococcus damnosus and other contaminating microbes will help prevent the spoilage of produced beverages and stop the unnecessary loss of countless dollars for their companies. (Brodmann, 2011)
Lactic Acid Bacteria
Bacteria found among food spoilage and food-borne pathogens are abundant. Lactic acid bacteria (LAB) are found among Pediococcus damnosus. Experiments have proven that this bacterium is associated with both food and beverage spoilage. More so, Lactobacillus phage B1 genome has 97 percent identity with Pediococcus damnosus; ensuring that it affects spoilage just as lactic acid bacteria does (Marco, 2012).
Lactic Acid Bacteria In Wine
Lactic acid bacteria play an important role for wine quality. A species-specific multiplex PCR system was performed to detect the presence of lactic acid bacteria species in Pediococcus damnosus. DNA extraction, multiplex PCR and the analysis of results prove that LAB is present in this microbe. The strain shows extreme resistance to ethanol and other influences so they are likely to appear during or after malolactic fermentation (Petri, 2012).
Companies have benefited from researchers developing a rapid method that can be used for detection, quantification, and identification of this microorganism. GC-MS and other slow cultivation and microscopy techniques are being replaced by analytical methods and test kits that that only take 12 hours to complete (Brodmann, 2011). These new methods look for certain DNA sequences in order to determine the presence of contamination, and if so, how much and which species.
Figure 2-1: Wine fermentation
Some strains of microorganisms are capable of surviving and multiplying rapidly even after the fermentation and sulfating processes of wine production. There are strict microbiological regulations and qualifications that beverages must pass before being sold so contamination is a costly problem. Figure 2-1 shows the start of fermentation in different flavors of wine. Certain strains of Pediococcus damnosus can synthesize β-D-glucan, a glucose homopolymer that changes the viscosity of wine to be thicker than oil (Delaherche, 2004). These microorganisms used to be detected and quantified by selective counting on MRS plates, where the colony-growth step took seven days. Researchers have now developed a more time-efficient method (taking one day) using real-time quantitative PCR as a new technique that can be used for routine screening (Delaherche, 2004).
Laboratory Studies
Pediococcus damnosus produces exopolysaccharides that change the viscosity of and ruin products. Primers PF5 and PF6 have been analyzed via PCR, along with new primers PF1 and PF8 that target the dps gene (putatative glucan synthase gene). This gene was found vastly among isolated strains of both Pediococcus damnosus and Oenococcus oeni (Walling, 2005). In order to ascertain that the cells were indeed producing the wine-spoiling glucan, an aggregation test (binding test) was performed with Streptococcus pneumoniae type 37-specific antibodies. Agglutination proved that there is a direct relation between gene dps and glucan production (Walling, 2005). This shows that gene dps could be the cause of exopolysaccharide production not only in Pediococcus damnosus, but in Oenococcus oeni as well.
The microbe is further studied with different controls to determine resistance and sensitivities. Pediocin PD-1 is one, of many studied, bactericidal antimicrobial peptides. Pediococcus damnosus produces pediocin PD-1 and so researchers find this microbe to be timely. A gel electrophoresis experiment was used to detect pediocin PD-1 to be a molecular mass of 35 kDa. Researchers continued to undergo experiments with pediocin PD-1 determining the isoelectric point and antimicrobial activity. Results from their tests revealed that pediocin PD-1 is also resistant to specific treatments (Green, 1997).
While most microbes are harmless to humans Pedicococcus damnosus is yet undesired, specifically for beer and wine companies. It is considered a spoilage bacterium in wine making and brewing fields that also contributes to unwanted viscosity due to glucan synthesis. Researchers have supplied fast detection, quantification, and identification of this microbe to help the major distributing company’s preserve their products.
References
[1]Brodmann, Peter D, Fehlmann M, Gafner J, Methods and Kits Pertaining to the Detection, Identification and Quantification of Bacteria and Yeast in Wine, Beer, and Juices. ETS Laboratories, assignee. Patent US 8,017,359 B2. 13 Sept. 2011.
[2]Delaherche A, Claisse O, Lonvaud-Funel A, "Detection and Quantification of Brettanomyces bruxellensis and 'ropy' Pediococcus damnosus Strains in Wine by Real-time Polymerase Chain Reaction." Journal of Applied Microbiology 97.5 (2004): 910-15.
[3]Green G, Dicks L.M.T, Bruggeman G, Vanamme E.J, Chikindas M.L 1997. Pediocin PD-1, a bactericidal antimicrobial peptide from Pediococcus damnosus NCFB 1832. Journal of Applied Microbiology(83): 127-132.
[4]Marco M, Garneau J, Tremblay D, Quiberoni A, Moineau S 2012. Characterization of Two Virulent Phages of Lactobacillus plantarum. Applied and Environmental Microbiology (78):8719-8734.
[5]Petri A, Pfannebecker J, Frohlich J, Konig H 2012. Fast identification of wine related lactic acid bacteria by multiplex PCR. Food Microbiology (33): 48-54.
[6]Walling E,Gindreau E, Lounvaud-Funel A, "A Putative Glucan Synthase Gene dps Detected in Exopolysaccaride-Producing Pediococcus damnosus and Oenococcus oeni Strains Isolated From Wine and Cider." International Journal of Food Microbiology (2005): 53-62.