Photobacterium leiognathi: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
{{Uncurated}} | {{Uncurated}} | ||
{{Biorealm Genus}}''Photobacterium leiognathi'' | {{Biorealm Genus}}''Photobacterium leiognathi'' | ||
[[Image:Photobacterium leiognathi on agar plate.jpeg| | [[Image:Photobacterium leiognathi on agar plate.jpeg|350px|thumb|right|alt=alt|Figure 1. Plate culture of Photobacterium leiognathi. Courtesy of Pearson Educacion]] | ||
==Classification== | ==Classification== | ||
| Line 11: | Line 12: | ||
===Species=== | ===Species=== | ||
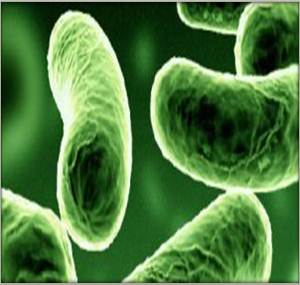
[[Image:Photobacterium leiognathi.jpg|frame|right|150px|alt=alt|Figure 2. Photobacterium leiognathi cells. Courtesy of ALL-Earth.org]] | |||
{| | {| | ||
| height="10" bgcolor="#FFDF95" | | | height="10" bgcolor="#FFDF95" | | ||
| Line 17: | Line 20: | ||
''Photobacterium leiognathi'' | ''Photobacterium leiognathi'' | ||
==Description and significance== | ==Description and significance== | ||
Photobacterium leiognathi is one of the many species within the genus Photobacterium and the family of Vibrionales. P. leiognathi is a gram negative, coccobacillus-straight shaped flagellated bacterium. P. leiognathi has no pigmentation and appears white or colorless unless found in colonies where it exhibits luminescent properties. P. leiognathi is a facultative anaerobic chemo-organothroph bacterium. P. leiognathi is a marine organism, which requires sodium for growth and lives as a free-living organism in warm waters or in association with fish. [8,10,11,12] | |||
==Genome structure== | ==Genome structure== | ||
two circular chromosomes | |||
highest level of assembly is scaffold only, currently 4,598,918 total base pair | |||
nucleotide sequence 7.3kbp | |||
lacks luxF gene, which is significant | |||
due to the 16S RNA many species, including this one have occasionally been shifted in and out of the genus | |||
some genomic polymorphism has been seen within the species, depending on the organism they form symbiotic relationships with, currently 11 subspecies of P. leiognathi are known | |||
==Cell and colony structure== | ==Cell and colony structure== | ||
gram negative | |||
coccobacillus (plump or straight rod) | |||
diameter 0.8-1.3µm and length 1.8-2.4µm | |||
1-3 flagella, Some P.leiognathi subspecies which have formed symbiotic relationship with fish have been discovered to have no flagella. | |||
Individual cells are colorless | |||
white or colorless – exhibit bioluminescense | |||
can emit bluish-green light (490nm) | |||
lux genes – involved in oxidation of a reduced riboflavin mononucleotide (FMNH2) and a long chain aldehyde | |||
==Metabolism== | ==Metabolism== | ||
facultative anaerobe | |||
can perform fermentative and respiratory functions | |||
chemo-organotroph | |||
glucose, mannose, fructose, or glycerol as sources of carbon | |||
requires sodium for growth | |||
==Ecology== | ==Ecology== | ||
P. leiognathi might be found as a free living marine organism in tropical waters. P. leiognathi grows best in water temperatures between 18-25̊ C around the mesopelagic zone (200-1000m depth). P. leiognathi can also form symbiotic relationships with fish and squid, as part of their light organs. P. leiognathi can also act as a decomposer of dead fish. | |||
P. leiognathi uses its bioluminescence to attract predators | |||
==Pathology== | ==Pathology== | ||
Other species of photobacteria are pathogenic, P. leiognathi is not even though it has the same ToxR transmembrane binding protein often associated with gastroenteritis caused by seafood consumption in mammals | |||
==References== | ==References== | ||
[1] Nijvipakul, Sarayut, et al. 2008, "LuxG Is a Functioning Flavin Reductase for Bacterial Luminescence", American Society for Microbiology: Journal of Bacteriology, Vol 190, No 5, pg. 1531-1538,doi: 10.1128/JB.01660-07 | [1] Nijvipakul, Sarayut, et al. 2008, "LuxG Is a Functioning Flavin Reductase for Bacterial Luminescence", American Society for Microbiology: Journal of Bacteriology, Vol 190, No 5, pg. 1531-1538,doi: 10.1128/JB.01660-07 | ||
[2] Valentine N. Petushkov, Bruce G. Gibson,and John Lee. 1995, “Properties of recombinant fluorescent proteins from Photobacterium leiognathi and their interaction with luciferase intermediates”, Biochemistry including biophysical chemistry and molecular biology. Vol 34, No 10, pg.3300-3309, DOI: 10.1021/bi00010a020 | [2] Valentine N. Petushkov, Bruce G. Gibson,and John Lee. 1995, “Properties of recombinant fluorescent proteins from Photobacterium leiognathi and their interaction with luciferase intermediates”, Biochemistry including biophysical chemistry and molecular biology. Vol 34, No 10, pg.3300-3309, DOI: 10.1021/bi00010a020 | ||
[3] Lee,Chan Yong, Rose B. Szittner, and Edward A. Meighen. 1991, “The lux genes of the luminous bacterial symbiont, Photobacterium leiognathi, of the ponyfish; nucleotide sequence, difference in gene organization, and high expression in mutant Escherichia coli” European Journal of Biochemistry, Vol 201, Issue 1, pg. 161-167, DOI: 10.1111/j.1432-1033.1991.tb16269.x | [3] Lee,Chan Yong, Rose B. Szittner, and Edward A. Meighen. 1991, “The lux genes of the luminous bacterial symbiont, Photobacterium leiognathi, of the ponyfish; nucleotide sequence, difference in gene organization, and high expression in mutant Escherichia coli” European Journal of Biochemistry, Vol 201, Issue 1, pg. 161-167, DOI: 10.1111/j.1432-1033.1991.tb16269.x | ||
[4] Herring, Peter. 2002, “Marine microlights: the luminous marine bacteria” Microbiology Today, Vol 29, pg. 174-176 | [4] Herring, Peter. 2002, “Marine microlights: the luminous marine bacteria” Microbiology Today, Vol 29, pg. 174-176 | ||
[5] Coil, David. 2011, “Microbiology Christmas Tree – luminescent bacteria,giant microbes, and more” Microbiology of the Built Environment Network | [5] Coil, David. 2011, “Microbiology Christmas Tree – luminescent bacteria,giant microbes, and more” Microbiology of the Built Environment Network | ||
[6] Boisvert, H. 1967, “Photobacterium leiognathi” National Center for Biotechnology Information | [6] Boisvert, H. 1967, “Photobacterium leiognathi” National Center for Biotechnology Information | ||
[7] Yirka, Bob. 2011, “Research shows ocean bacteria glow to attract those that would eat them” Physorg.com. doi: 10.1073/pnas.1116683109 | [7] Yirka, Bob. 2011, “Research shows ocean bacteria glow to attract those that would eat them” Physorg.com. doi: 10.1073/pnas.1116683109 | ||
Revision as of 05:24, 4 May 2012
A Microbial Biorealm page on the genus Photobacterium leiognathi Photobacterium leiognathi
Classification
Higher order taxa
Domain (Bacteria); Phylum (Proteobacteria); Class (Gammaproteobacteria); Order (Vibrionales); Family (Vibrionales); Genus (Photobacterium)
Species
|
NCBI: Taxonomy |
Photobacterium leiognathi
Description and significance
Photobacterium leiognathi is one of the many species within the genus Photobacterium and the family of Vibrionales. P. leiognathi is a gram negative, coccobacillus-straight shaped flagellated bacterium. P. leiognathi has no pigmentation and appears white or colorless unless found in colonies where it exhibits luminescent properties. P. leiognathi is a facultative anaerobic chemo-organothroph bacterium. P. leiognathi is a marine organism, which requires sodium for growth and lives as a free-living organism in warm waters or in association with fish. [8,10,11,12]
Genome structure
two circular chromosomes highest level of assembly is scaffold only, currently 4,598,918 total base pair nucleotide sequence 7.3kbp lacks luxF gene, which is significant due to the 16S RNA many species, including this one have occasionally been shifted in and out of the genus some genomic polymorphism has been seen within the species, depending on the organism they form symbiotic relationships with, currently 11 subspecies of P. leiognathi are known
Cell and colony structure
gram negative coccobacillus (plump or straight rod) diameter 0.8-1.3µm and length 1.8-2.4µm 1-3 flagella, Some P.leiognathi subspecies which have formed symbiotic relationship with fish have been discovered to have no flagella. Individual cells are colorless white or colorless – exhibit bioluminescense can emit bluish-green light (490nm) lux genes – involved in oxidation of a reduced riboflavin mononucleotide (FMNH2) and a long chain aldehyde
Metabolism
facultative anaerobe can perform fermentative and respiratory functions chemo-organotroph glucose, mannose, fructose, or glycerol as sources of carbon requires sodium for growth
Ecology
P. leiognathi might be found as a free living marine organism in tropical waters. P. leiognathi grows best in water temperatures between 18-25̊ C around the mesopelagic zone (200-1000m depth). P. leiognathi can also form symbiotic relationships with fish and squid, as part of their light organs. P. leiognathi can also act as a decomposer of dead fish. P. leiognathi uses its bioluminescence to attract predators
Pathology
Other species of photobacteria are pathogenic, P. leiognathi is not even though it has the same ToxR transmembrane binding protein often associated with gastroenteritis caused by seafood consumption in mammals
References
[1] Nijvipakul, Sarayut, et al. 2008, "LuxG Is a Functioning Flavin Reductase for Bacterial Luminescence", American Society for Microbiology: Journal of Bacteriology, Vol 190, No 5, pg. 1531-1538,doi: 10.1128/JB.01660-07
[2] Valentine N. Petushkov, Bruce G. Gibson,and John Lee. 1995, “Properties of recombinant fluorescent proteins from Photobacterium leiognathi and their interaction with luciferase intermediates”, Biochemistry including biophysical chemistry and molecular biology. Vol 34, No 10, pg.3300-3309, DOI: 10.1021/bi00010a020
[3] Lee,Chan Yong, Rose B. Szittner, and Edward A. Meighen. 1991, “The lux genes of the luminous bacterial symbiont, Photobacterium leiognathi, of the ponyfish; nucleotide sequence, difference in gene organization, and high expression in mutant Escherichia coli” European Journal of Biochemistry, Vol 201, Issue 1, pg. 161-167, DOI: 10.1111/j.1432-1033.1991.tb16269.x
[4] Herring, Peter. 2002, “Marine microlights: the luminous marine bacteria” Microbiology Today, Vol 29, pg. 174-176
[5] Coil, David. 2011, “Microbiology Christmas Tree – luminescent bacteria,giant microbes, and more” Microbiology of the Built Environment Network
[6] Boisvert, H. 1967, “Photobacterium leiognathi” National Center for Biotechnology Information
[7] Yirka, Bob. 2011, “Research shows ocean bacteria glow to attract those that would eat them” Physorg.com. doi: 10.1073/pnas.1116683109
Edited by Jossary Gerry, student of Dr. Lisa R. Moore, University of Southern Maine