Pirellula: Difference between revisions
No edit summary |
|||
| (3 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
{ | {{Curated}} | ||
{{Biorealm Genus}} | |||
[[Image:pirellula_and_flagella.JPG|thumb|250px|right|Electron micrograph of a ''Pirellula'' bacterium from giant tiger prawn tissue (''Penaeus monodon''). Notice the large crateriform structures (C) on the cell surface and flagella. From [http://www.pubmedcentral.gov/articlerender.fcgi?tool=pubmed&pubmedid=1781677 Fuerst'' et al.'']]] | [[Image:pirellula_and_flagella.JPG|thumb|250px|right|Electron micrograph of a ''Pirellula'' bacterium from giant tiger prawn tissue (''Penaeus monodon''). Notice the large crateriform structures (C) on the cell surface and flagella. From [http://www.pubmedcentral.gov/articlerender.fcgi?tool=pubmed&pubmedid=1781677 Fuerst'' et al.'']]] | ||
| Line 14: | Line 13: | ||
''Pirellula marina, P. staleyi, P. sp.'' | ''Pirellula marina, P. staleyi, P. sp.'' | ||
{| | |||
| height="10" bgcolor="#FFDF95" | | |||
'''NCBI: [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=123 Taxonomy] [http://www.ncbi.nlm.nih.gov/genomes/framik.cgi?db=Genome&gi=308 Genome]''' | |||
|} | |||
==Description and Significance== | ==Description and Significance== | ||
| Line 28: | Line 32: | ||
[[Image:fimbrae1.JPG|thumb|300px|left|Electron micrograph of a ''Pirellula'' bacterium showing the large crateriform structures (c) and the fimbriae (f) on the cell pole. From [http://www.pubmedcentral.gov/articlerender.fcgi?tool=pubmed&pubmedid=1781677 Fuerst'' et al.'']]] | [[Image:fimbrae1.JPG|thumb|300px|left|Electron micrograph of a ''Pirellula'' bacterium showing the large crateriform structures (c) and the fimbriae (f) on the cell pole. From [http://www.pubmedcentral.gov/articlerender.fcgi?tool=pubmed&pubmedid=1781677 Fuerst'' et al.'']]] | ||
''Pirellula'' sp. strain 1 ("''Rhodopirellula baltica''") is a marine, aerobic, heterotrophic bacterium. It has all the genes for a functional flagellum (and only one essential gene for chemotaxis). Like all other Planctomycetes, ''Pirellula'' sp. strain 1 reproduces by budding in which a stalk formation produces aggregates that form into motile daughter cells. Also characteristic of Planctomycetes, ''Pirelulla'' bacterial cell walls contain no peptidoglycan; in fact, Planctomycetes are the only group of free-living bacteria to lack peptidoglycan. The cell walls are instead stabilized by the protein sacculus with disulfide bonds. The presence of several genes encoding for some of the steps in peptidoglycan biosynthesis leads some to the conclusion that Planctomycetes evolved after peptidoglycan was around; it is suggested that after the Planctomycetes developed a proteinaceous cell envelope, they lost some of the genes necessary for peptidoglycan biosynthesis. It is also suggested that the Planctomycetes evolved from Gram-negative cells due to the presence of a few genes encoding a few of the enzymes necessary for Gram-negative membranes in the ''Pirellula'' sp. strain 1 genome (Glockner ''et al.'' 2003). Another characteristic property of ''Pirellula'' and other Planctomycetes are their complex internal structures such as an intracytoplasmic membrane surrounding the nuclear regions and compartmentalizing the cell as well as ribosomes located only within the intracytoplasmic membrane. However, unlike ''Gemmata'' bacteria, ''Pirellula'' ''marina'' and ''Pirellula'' ''staleyi'' have only a single enveloping membrane while ''Gemmata'' ''obscuriglobus'' has a double membrane (2). (See [http://biology.kenyon.edu/Microbial_Biorealm/bacteria/gemmata/Gemmata.htm Gemmata].) | |||
[[Image:pirellula_budding.JPG|thumb|200px|left|Electron mircrograph of a ''Pirellula'' bacterium displaying a mother cell with a bud. Notice the large crateriform structures on the reproductive pole of the mother cell and over half of the bud. Small crateriform structures exist is clusters on poles opposite to the concentrated areas of the large crateriform structure. Also notice the fimbriae projecting from the mother cell. Bar = 0.5 microliters. From [http://www.pubmedcentral.gov/articlerender.fcgi?tool=pubmed&pubmedid=1781677 Fuerst'' et al.'']]] | [[Image:pirellula_budding.JPG|thumb|200px|left|Electron mircrograph of a ''Pirellula'' bacterium displaying a mother cell with a bud. Notice the large crateriform structures on the reproductive pole of the mother cell and over half of the bud. Small crateriform structures exist is clusters on poles opposite to the concentrated areas of the large crateriform structure. Also notice the fimbriae projecting from the mother cell. Bar = 0.5 microliters. From [http://www.pubmedcentral.gov/articlerender.fcgi?tool=pubmed&pubmedid=1781677 Fuerst'' et al.'']]] | ||
The genome ''Pirellula'' sp. strain 1 has two orders of magnitude more sulfatase genes than those in other known prokaryotic genomes: it includes 110 genes that encode for sulfatase proteins and funcitons, 82 (75%) of which are similar to prokaryotic sulfatases and 28 (25%) of which are similar to eukaryotic sulfatases. Bacterial sulfatases are generally used in sulfur scavenging and are regulated by sensing sulfur deprivation. However, marine systems in which Pirellula generally live usually have high inorganic sulfate concentrations; thus, the sulfatases might be used for the function of accessing "more effectively the carbon skeleton of sulfated compounds as an energy source rather than to meet its sulfate requirements" (Glockner ''et al.'' 2003). | The genome ''Pirellula'' sp. strain 1 has two orders of magnitude more sulfatase genes than those in other known prokaryotic genomes: it includes 110 genes that encode for sulfatase proteins and funcitons, 82 (75%) of which are similar to prokaryotic sulfatases and 28 (25%) of which are similar to eukaryotic sulfatases. Bacterial sulfatases are generally used in sulfur scavenging and are regulated by sensing sulfur deprivation. However, marine systems in which Pirellula generally live usually have high inorganic sulfate concentrations; thus, the sulfatases might be used for the function of accessing "more effectively the carbon skeleton of sulfated compounds as an energy source rather than to meet its sulfate requirements" (Glockner ''et al.'' 2003). | ||
Latest revision as of 20:35, 6 August 2010
A Microbial Biorealm page on the genus Pirellula
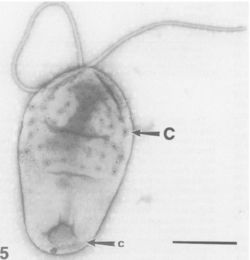
Classification
Higher order taxa:
Bacteria; Planctomycetes; Planctomycetacia; Planctomycetales; Planctomycetaceae
Species:
Pirellula marina, P. staleyi, P. sp.
Description and Significance
Pirellula sp. strain 1, which as also been called Rhodopirellula baltica, is a marine bacterium that can be found all over the world.
Genome Structure
Pirellula sp. strain 1 ("Rhodopirellula baltica") has a circular genome comprised of 7,145, 576 bases, which makes it the largest prokaryotic circular genome known so far. 1,301 of the genes (which is 17.6% of all genes) that encoded for an average length of 464 amino acids have multiple copies in the genome - in fact, multicopy genes make up about 25.4% of the entire genome sequence. This number is still less than the known 30% in Bacillus subtilis and the calculated 29% in the genome of E. coli. The G-C content of the genome of Pirellula sp. strain 1 is 55.4 mol%. (Glockner et al. 2003). It is thought that because Pirellula sp. strain 1 contains genes similar to archea-like genes. While there are speculations about why this occured the most common theory is that there was some lateral gene transfer from archea to a ancestor of Pirellula. This seems to be more likely than Pirellula retaining a gene from a common ancestor between archea and bacteria.
Cell Structure and Metabolism
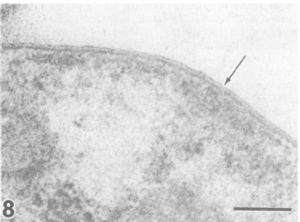
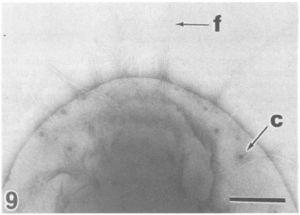
Pirellula sp. strain 1 ("Rhodopirellula baltica") is a marine, aerobic, heterotrophic bacterium. It has all the genes for a functional flagellum (and only one essential gene for chemotaxis). Like all other Planctomycetes, Pirellula sp. strain 1 reproduces by budding in which a stalk formation produces aggregates that form into motile daughter cells. Also characteristic of Planctomycetes, Pirelulla bacterial cell walls contain no peptidoglycan; in fact, Planctomycetes are the only group of free-living bacteria to lack peptidoglycan. The cell walls are instead stabilized by the protein sacculus with disulfide bonds. The presence of several genes encoding for some of the steps in peptidoglycan biosynthesis leads some to the conclusion that Planctomycetes evolved after peptidoglycan was around; it is suggested that after the Planctomycetes developed a proteinaceous cell envelope, they lost some of the genes necessary for peptidoglycan biosynthesis. It is also suggested that the Planctomycetes evolved from Gram-negative cells due to the presence of a few genes encoding a few of the enzymes necessary for Gram-negative membranes in the Pirellula sp. strain 1 genome (Glockner et al. 2003). Another characteristic property of Pirellula and other Planctomycetes are their complex internal structures such as an intracytoplasmic membrane surrounding the nuclear regions and compartmentalizing the cell as well as ribosomes located only within the intracytoplasmic membrane. However, unlike Gemmata bacteria, Pirellula marina and Pirellula staleyi have only a single enveloping membrane while Gemmata obscuriglobus has a double membrane (2). (See Gemmata.)
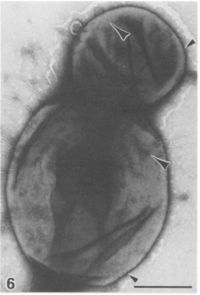
The genome Pirellula sp. strain 1 has two orders of magnitude more sulfatase genes than those in other known prokaryotic genomes: it includes 110 genes that encode for sulfatase proteins and funcitons, 82 (75%) of which are similar to prokaryotic sulfatases and 28 (25%) of which are similar to eukaryotic sulfatases. Bacterial sulfatases are generally used in sulfur scavenging and are regulated by sensing sulfur deprivation. However, marine systems in which Pirellula generally live usually have high inorganic sulfate concentrations; thus, the sulfatases might be used for the function of accessing "more effectively the carbon skeleton of sulfated compounds as an energy source rather than to meet its sulfate requirements" (Glockner et al. 2003).
Ecology
Although Pirellula is globally distributed marine bacteria, planctomycetes, in general, can be found abundantly in terrestrial and marine habitats alike. For example, a study of the planctomycetes make-up of anoxic and aerobic basins and an anaerobic digestor at a wastewater treatment plant found the samples to be 32% Pirellula.
