Pseudomonas Aeruginosa Infection and Biofilm Production in Immunocompromised Individuals: Difference between revisions
No edit summary |
|||
| (24 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
{{Uncurated}} | |||
[[Image:P.jpg|thumb|300px|right| Electron microscopy of <i>P. aeruginosa</i> http://www.pseudomonas.com/p_aerug.jsp.]] | |||
==Introduction== | |||
<br> <i>P. aeruginosa</i> is a gram negative bacterium with one flagella, as can be seen in Figure 1 [[#References|[10]]]. The flagellum plays an extremely important role in the production and assembly of biofilm formation. <i>P. aeruginosa</i> is also a facultative anaerobe, but its optimal metabolism is aerobic respiration. In the absence of oxygen, it can respire anaerobically using nitrate or an alternate electron acceptor [[#References|[4]]]. However, it often cannot grow when it is simultaneously a pathogen and not exposed to oxygen, as is common in the depth of the biofilm. Many cells thus often remain in an un-dividing state when in a biofilm. This, however, works in <i>P. aeruginosa’s</i> favor, as many antibiotics cannot kill cells that are not actively growing or dividing [[#References|[16]]]. Furthermore, it can catabolize a variety of organic molecules, even carcinogens such as benzoate. This incredible nutritional and metabolic versatility and diversity allows <i>P. aeruginosa</i> to survive attack by both phage and human induced antimicrobial agents, as well as live in a diverse array of environments. <i>P. aeruginosa</i> is found ubiquitously, in environments as diverse as hospitals, sewage, animals, plants, water, and the soil. It is also the most predominant organism in oligotrophic aquatic ecosystems. Furthermore, in the soil, <i>P. aeruginosa</i> is an important ecological bacterial species, breaking down the polycyclic aromatic hydrocarbons, and catabolizing important molecules such as rhamnolipids, quinolones, and hydrogen cyanide [[#References|[10]]].This versatility allows it to be the most abundant organism on earth. <br> | |||
<br> Despite this abundance, it is an opportunistic pathogen, rarely infecting healthy individuals. It is common in immunocompromised patients, such as people suffering from Acquired Immunodeficiency Syndrome (AIDS) or cancer, as well as burn victim (Davis, 2003; Lederberg, 2000). Additionally, many Cystic Fibrosis (CF) patients are chronically infected by <i>P. aeruginosa</i>, causing severe pulmonary inflammation and damage. There are many hypotheses explaining the abundance of <i>P. aeruginosa</i> infections in the CF population, but the most well supported is the high-salt hypotheis, which states that high rate of infection are due to CF patients having an abundance of salt in their respiratory tract; this prevents the production and action of chemicals and proteins used by the immune system to prevent infection [[#References|[5]]] [[#References|[16]]]. Because of its prevalence in hospitals and selective pathogenesis, <i>P. aeruginosa</i> infects two thirds of critically ill hospitalized patients. <br> | |||
<br>Unfortunately, it is an extremely invasive pathogen, with a mortality rate of 40-60%. The mortality rate is extremely high because of the biofilm nature of <i>P. aeruginosa</i>. When a pathogen is present in the human lungs, it often forms biofilms, communities of bacterial cells attached to an inert or living surface and enclosed in an extracellular polysaccharide matrixes. These biofilms release planktonic bacteria, which disperse to spread the infection to other tissues or host organisms. Many antibiotics kill the planktonic bacteria. However, antibiotics fail to kill the bacterium in the biofilm; for this reason, after antibiotic therapy is complete, the biofilm continues to produce planktonic bacteria and the infection reoccurs. Biofilms are inherently resistant to antibiotics, whether in the form of bacteriophages, amoebae, or chemical biocides [[#References|[10]]]. | |||
==Pathology== | |||
[[Image:CFF.jpg|thumb|300px|right| (A) Explanted lung of a CF patients. Green mucus is spilling out of the major airways. (B) A CF lung that has been fixed by formalin. http://www.ncbi.nlm.nih.gov/pubmed/19418571.]] | |||
<br><u><b> Clinical Feature</b></u><br> | |||
<i>P. Aeruginosa</i> can infect may parts of the body. It is most often seen, however, in the respiratory tract due to its prevalence in the lower respiratory tract of the CF population. Here it generally causes pneumonia, which has symptoms such as fever, cyanosis, the chills, and a productive cough. It is however also seen in the blood, which is usually acquired from medical devices. It may also be acquired from native and prosthetic heart valves, causing endocarditis. Causing meningitis and brain abscesses, it also infects the Central Nervous System. This too, is usually the result of a medical procedure, such as paranasal sinus surgery. When infecting the eye, <i>P. aeruginosa</i>, produces extracellular enzymes that cause rapid destruction of the infected lesion. This causes many forms of vision impairment, such as bacterial keratitis, scleral abscesses, and ophthalmic neonatorum. It can also be found in the ear, skin, gastrointestinal tract, urinary tract, and bones [[#References|[20]]]. <br> | |||
<b><u>Transmission and Diagnosis</u></b><br> | |||
<i>P. aeruginosa</i> is an ubiquitous organism, found in diverse environments, such as hospitals, sewage, animals, plants, water, and the soil (Lederberg, 2000). It is also the most predominant organism in oligotrophic aquatic ecosystems. Despite, this abundance, <i>P. aeruginosa</i> is an opportunistic pathogen, rarely infecting healthy individuals (Stewart, 2001). Infections occur in patients with compromised immune systems, such as people with acquired immune deficiency syndrome (AIDS). They also occur when the bacterium surpass the physical barriers of the innate immune system, which can be seen in the high rate of infections in burn victims [[#References|[5]]]. In most cases, <i>P. aeruginosa</i> infections spread though contact with other, infected individuals. One study by Johnson and colleagues used gel electrophoresis to find that in a selected Intensive Care Unit, 31% of Imipenem resistant <i>P. aeruginosa</i> cases were due to patient-to-patient transmission. Even more interestingly, 19% of cases were from the patients’ own endogenous flora [[#References|[21]]]. <br> | |||
<br> The diagnosis of <i>P.aeruginosa</i> infections can be made several ways based on its identifying features. The bacterium grows well on laboratory media, especially eosin-methylthionine blue agar and blood agar. It can be identified based on it Gram negative morphology, fruity odor, and ability to grow at high temperature, like 42 degrees C. Due to the chemical pyocyanin, it can florescence blue under ultraviolet light. This helps with its early identification in wounds [[#References|[18]]]. Finally, real time Polymerase Chain Reaction (PCR) of cough and septum swabs is a new, exciting way to quickly and accurately detect the presence of <i>P. aeruginosa</i> [[#References|[22]]]. <br> | |||
<br> Once infected with <i>P. aeruginosa</i>, the prognosis is poor: it has a mortality rate of 40-60%. This mortality rate is due to the biofilm nature of <i>P. aeruginosa</i>. When a pathogen colonizes human lungs, it often forms biofilms, communities of bacterial cells attached to an inert or living surface and enclosed in an extracellular polysaccharide matrixes. These biofilms release planktonic bacteria, which are dispersed to spread the infection to other tissues or host organism. Many antibiotics kill the planktonic bacteria. However, antibiotics fail to kill the bacterium in the biofilm; for this reason, after antibiotic therapy is complete, the biofilm continues to produce planktonic bacteria and the infection reoccurs [[#References|[10]]]. This reoccurring infection results slowly destroys the infected organs through the release of toxins. <br> | |||
<br> <u>In Burn Victims</u><br> | |||
<i>P. aeruginosa</i> is the most common form of infection suffered by burn victims. Human skin is comprised of two layers, the epidermal and dermis. When the skin is burned, it allows microbes to colonize the subcutaneous tissue. In an experiment done by Trafny and colleagues, animals with partial-thickness cutaneous burns developed mature biofilms in 48 to 72 hours [[#References|[17]]]. <i>P.aeruginosa</i> biofilms have been shown to be fully mature in humans only 10 hours after burns [[#References|[9]]]. <br> | |||
<br><u>In Cystic Fibrosis (CF) patients</u> | |||
[[Image:F.jpg|thumb|300px|right| The major hypothesis causing <i>P.aeruginosa</i> infections in CF patiens. http://www.sciencedirect.com/science/article/pii/S1526055002000033.]] | |||
CF is an inherited autosomal recessive disease that results from mutations in the gene encoding the CF transmembrane conductance regulator (CFTR) protein.<br> CFTR is a bidirectional chloride channel and unidirectional sodium channel in epithelial cells [[#References|[5]]]. If this protein is not fully functional, chloride cannot move into and out of the cells and sodium cannot cross the cell membrane; as chloride is important for water movement, mucus becomes thick. <br> In healthy lungs, entering <i>P. aeruginosa</i> are systematically removed or killed quickly by a variety of innate immune responses, such as mucociliary clearance and macrophages. However, when CF patients contract <i>P. aeruginosa</i>, it causes chronic infection. 85% of CF patients have chronic [[#References|[5]]]<i>P. aeruginosa</i> infections by adolescence. This elicits an aggressive inflammatory response, which causes lung damage. There are currently several hypotheses explaining the prevalence of <i>P. aeruginosa</i>, which are diagramed in Figure 3. [[#References|[7]]]<br> | |||
<br><i>Decreased Clearance of Mucociliary Passages</i><br> | |||
To begin, some propose that the sodium and water hyperabsorption in CF causes the airway surface liquid (ASL) and mucus layer to have an abnormally small volume. This causes the cilia to beat inefficiently, impairing the ability of CF patients to clear their mucociliary passages (throat). This causes inhaled pathogens to become trapped, promoting inflammation and infection [[#References|[5]]]. Experiments have shown that treatment with hypertonic saline improves lung function and mucus clearance rate, which could reduce the occurrence of bacterial entrapment causing <i>P. aeruginosa</i> infection [[#References|[7]]].<br> | |||
<br><i>High Salt Deactivation Hypothesis</i><br> | |||
A second hypothesis centers on the high levels of sodium and chloride in the ASL. Many immune defenses, such as hBD-1 lysozymes and lactoferrin, are salt sensitive. Smith and colleagues showed that many of these defensives are rendered nonfunctional in high salt environments, like CF lungs, using an in vitro experiment [[#References|[15]]]. Another recent experiment by Bals and colleagues showed that hBD-2, a peptide with antimicrobial properties expressed throughout the epithelium, is sensitive to salt concentrations; its ability to inhibit bacterial growth diminished as the concentration of NaCl increased. Its ability to function in salt was tested by exposing <i>E. coli</i> cells to isolated hBD-2 peptide at differing NaCl concentration [[#References|[2]]]. This suggests that it would be nonfunctional in CF and this breach in the innate immunity could be the cause of many infections. <br> | |||
<br><i>Acidic Environment Hypothesis</i><br> | |||
Another possibility is that the acidic pH of CF epithelium impairs mechanisms of the immune systems. CFTR is involved in the transport of bicarbonate ions, which regulate cell surface pH, making CF epithelium abnormally acidic. This may stop immune system mechanisms, such as the mucociliary clearance or phagocyte function from fully working [[#References|[5]]]. While the decrease in ASL pH has not been shown conclusively in humans, Pezzulo and colleagues discovered that pigs lacking CFTR have more acidic ASL compared to pigs with functional CFTR [[#References|[13]]]. Furthermore, IL-8 is secreted in response to <i>P. aeruginosa</i> and causes severe inflammation. Its secretion, however, decreased with treatments of substances that inhibit acidification of intracellular organelles such as chloroquine. This shows that either <i>P. aeruginosa</i> is less effective in basic conditions, which is unlikely considering the range of environments it resides in, or that the immune system is more inept at less acidic conditions [[#References|[6]]]. <br> | |||
<br><i>aGMI Receptor Hypothesis</i><br> | |||
<i>P. aeruginosa</i> used several different types of organelles, including pili and flagella for adherance to cell surface receptors. The receptor for both pili and flagella is GalNAcβ1-4Gal, which consists of several glycolipids. Studies, such as one done by Saiman and colleagues, has shown that one of the glycoproteins, asialoGM1 (aGM1), is present in a higher concentration on CF epithelial cells compared to normal cells (12% v 2.9% , respectively) [[#References|[14]]].<br> | |||
<br><i>Inflammation Hypothesis</i><br> | |||
Finally, research has shown that when <i>P. aeruginosa</i> DNA is released from the bacterium, it causes inflammation of CF epithelial cells. It does this by using p38 and Erk mitogen-activated protein kinase to induce the secretion of interleukin-8 (IL-8) from the host cells. This chemokine causes excessive neutrophil infiltration. This was determined in vitro by exposing bronchial epithelial cell lines with and without functional CTFR to <i>P. aeruginosa</i> and then quantifying the gene and protein products secreted [[#References|[6]]]. This research indicates that <i>P. aeruginosa</i> could cause inflammation of the epithelial cells without the bacteria even adhering to the cell surface [[#References|[5]]]. <br> | |||
<br>Tissues near the infected biofilm often become severely damages in CF patients due to the excessive amount of neutrophils and immune complexes, such as IL-8, released, as well as the toxins produced by the <i>P. aeruginos</i> a bacterium [[#References|[16]]]. This can be seen in Figure 2: the lungs are incredibly damaged. Because of the damage to pulmonary tissue, immunosuppression drugs are often given to CF patients— the innate immune system cannot fight off the biofilms and doing so just worsens health conditions. However, consistent treatment with antibiotics to prevent colonization in young patients is promising [[#References|[4]]]. <br> | |||
<br><u> In Hospitals</u><br> | |||
Due to the high presence of immunocompromised patients, <i>P. aeruginosa</i> infections are very common in hospitals. It infects two thirds of critically ill hospitalized patients [[#References|[10]]]. Much of the transmission is from biofilms that have colonized on medical devices. Pathogenic biofilms have been found using electron microscopy on the surface of medical devices, such as central venous catheters, prosthetic heart valves, and orthopedic implants (Costerton, 1999; Stewart, 2001). However, other studies, such as that done by Johnson and colleagues in a selected Intensive Care Unit showed that 31% of Imipenem resistant <i>P. aeruginosa</i> cases were due to patient-to-patient transmission. Even more interesting, 19% of cases were from the patients’ own endogenous flora [[#References|[21]]]. <br> | |||
==Biofilms== | ==Biofilms== | ||
[[Image:Bio.jpg|thumb|300px|. The formation of biofilms: highlighted are the attachment of planktonic cells, their multiplication and differentiation, and the detachment of newly formed planktonic cells. http://www.ncbi.nlm.nih.gov/pubmed/10334980. ]] | |||
Biofilms | Biofilms are organized communities of bacterial cells attached to an inert or living surface and enclosed in an extracellular polysaccharide matrix. This matrix forms a slippery, solid coat around the bacterial community used to protect the bacterial community in a hostile environment, such as the human lungs, where pathogens are systematically removed by a variety of innate immune responses, such as mucociliary clearance [[#References|[5]]]. These bacterial communities cause many persistent and chronic bacterial infections, such as <i>P. aeruginosa</i> infection in immunocompromised patients. An understanding of both biofilm production and interaction, therefore, is important because it could provide new therapeutic targets [[#References|[4]]]. <br> | ||
<br><b><u>Life Cycle of <i>P. aeruginosa</i></u></b><br> | |||
<u>Bacterial Attachment</u><br> | |||
<br>The initial attachment requires both flagella, as show by an experiment done by O’Toole and Kolter, where knock out mutants without flagella could not adequately adhere to a surface. During this attachment phase, the transcription of particular genes is activated, such as algC, algD, and algU, which are required for the synthesis of the extracellular polysaccharide that encases the biofilm. This is spectacular because it implies that <i>P. aeruginosa</i> have a ‘sense of touch’ that enables the detection and reaction to a surface [[#References|[4]]]. <br> | |||
<br><u> Formation of Microcolonies</u> <br> | |||
Next, microcolonies are formed. Microcolonies are relatively small groups of bacteria. The formation of microcolonies requires Type IV pili, as shown by O’Toole and Kolter. Bacteria lacking pili were able to form a monolayer, but not microcolonies. These pili are involved in a ‘twitching,’ which is thought to be necessary to the aggregation of cells into microcolonies [[#References|[4]]]. <br> | |||
<br><u>Differentiation</u><br> | |||
These microcolonies then differentiate into true biofilms. Quorum sensing, the exchange of chemical signals between cells, may affect the timing of this differentiation as well as the coordinated building of the biofilm into complex structures [[#References|[4]]]. <br> | |||
<br><u>Bloodstream Dissemination</u><br> | |||
Finally, the planktonic cells detach and differentiate from the biofilms. One mechanism for this to occur is that the pieces of biofilms break off, but this break may be regulated based on molecular cues; there appears to be a programmed detachment of planktonic bacteria. For example, it is thought that the release of planktonic cells requires an enzyme to digest alginate [[#References|[4]]]. Figure 4 outlines this entire process.<br> | |||
<br><b><u>Requirements for Biofilm Formation</u></b><br> | |||
<br><u>Gene Expression</u><br> | |||
<i>P. aeruginosa</i> planktonic cells and biofilm cells have extremely different lifestyles. Planktonic cells are not surrounded by an extracellular polysaccharide matrix, like their biofilm enclosed counterparts, but instead dissociate and diffuse from the biofilm in order to spread the bacteria to new tissues and host organisms. Because of this, it is surprising that the two types of <i>P. aeruginosa</i> cells differ only by 1% in terms of differential gene expression. This refutes the argument that <i>P. aeruginosa</i>’s existence in a biofilm is dependent on dramatic differences in gene expression. However, the small amount of genes that are activated and repressed (34 and 39 respectively) may be crucial for the aggregation of the cells into a biofilm. <br> | |||
<br> Gene specifying pili and flagella are suppressed in biofilm bacteria, suggesting that they are not needed for the incorporation and upkeep of a mature biofilm. They are instead probably important only in the initial process of biofilm development. Furthermore, the genes with the highest expression in the biofilm <i>P. aeruginosa</i> were those from a bacteriophage related to Pf1. Pf1 genes were activated 100 to 1000 times more in biofilms than planktonic bacteria. This shows that phage induction is important for gene transfer within biofilms. These genes are suspected to function either to exclude certain strains of <i>P. aeruginosa</i> from biofilms induction or to contain a gene specifying an important toxin [[#References|[19]]].<br> | |||
<br><u>Quorum Sensing</u><br> | |||
Quorum sensing is thought to determine when <i>P. aeruginosa</i> and other Gram Negative microcolonies differentiate into true biofilms. Individual cells produce acylhomoserine lactone signals; when a sufficient number of acylhomoserine producing cells are present, the signals accumulate and trigger the transcription and translation of a specific set of genes (Costerton, 1999). <br> | |||
<br> <i>LasR-LasI</i> is a gene that is also related to quorum sensing and differentiation in <i>P. aeruginosa</i>. It controls the expression of extracellular virulence factors, as well as the Rh1R and Rh1I, which control the expression of many secondary metabolites, such as butyrylhomoserine. Mutants of this process do not differentiate [[#References|[4]]]. <br> | |||
<br>It is believed that if cell-to-cell signaling is blocked or destroyed, biofilm production and maintenance would cease. If the biofilm were destroyed, it could allow the host immune system (or the host immune system supplemented with traditional antibiotics) to destroy the <i>P. aeruginosa</i> cells, in much the same way that antibiotics kill planktonic <i>P. aeruginosa</i> cells. [[#References|[5]]][[#References|[16]]]. <br> | |||
< | <br><u>Iron and Biofilm Production</u><br> | ||
P. aeruginosa | Iron is a signal for biofilm production in <i>P. aeruginosa</i>. It is believed that either the level of internal iron or the active transport of chelated iron acts as a signal to produce biofilm formation. Iron starvation can often prevent the growth of bacteria, but sufficient levels may serve as a signal for biofilm development. This has been demonstrated by Lactoferrin, which sequesters iron and prevents <i>P. aeruginosa</i> from maturing from thin monolayers into multicellular biofilm structures. This could be due to the fact that low iron causes continuous twitching movements, the control of which is needed to form biofilm structures. Iron is often acquired by pyverdine, which is regulated by <i>PvdS</i>. When pyverdine binds to an outer-membrane receptor, it results in a signal that governs not only the activity of <i>PvdS</i>, but extracellular virulence factors. This is important for the opportunistic nature of <i>P. aeruginosa</i> infections; only when iron concentration is high and the cell is tethered to a surface, can pathogenic biofilm production begin. The <i>Fur</i> protein also regulates iron-responsive genes through a pair of small regulatory RNAs. The authors found that <i>any</i> pathway that allows iron into the cell –pryverdine or <i>Fur</i>, for example— can function as an iron-signaling pathway (and thus as a signal for virulence and biofilm development pathway). This indicates that a critical content of intracellular iron is a signal for biofilm pathway initiation [[#References|[1]]]. <br> | ||
<br> | |||
==Antibiotic Resistance== | ==Antibiotic Resistance== | ||
[[Image:anti.jpg|thumb|300px|right| This figure shows the differeences in antibiotic resistance between mutator CF cells (black), nonmutator CF cells (grey) and non-CF cells (white). There are significant differences between the mutator and non-mutator CF cells when immersed in ticarcillin, ceftazidime, gentamicin, amikacin, norfloxacin, and fosfomycin, all antibiotics. | [[Image:anti.jpg|thumb|300px|right| This figure shows the differeences in antibiotic resistance between mutator CF cells (black), nonmutator CF cells (grey) and non-CF cells (white). There are significant differences between the mutator and non-mutator CF cells when immersed in ticarcillin, ceftazidime, gentamicin, amikacin, norfloxacin, and fosfomycin, all antibiotics. http://www.sciencemag.org/content/288/5469/1251.short.]] | ||
<br>Biofilms are inherently resistant to antibiotics, whether in the form of bacteriophages, amoebae, or chemical biocides. They have this resistance through multiple mechanisms; however, some antibiotics are more effective than others at penetrating and killing cells in the biofilm (figure 3) [[#References|[16]]]. <br> | |||
<br><b><u>Efflux Pumps</u></b><br> | |||
Often, bacteria avoid antibiotic-caused bactericide through the use of efflux pumps. However, these mutations are not the sole cause of antimicrobial protection in <i>P. aeruginosa</i> biofilms. <br> | |||
<br>Studies on genes encoding efflux pumps, for example, revealed that P. aeruginosa with multidrug efflux pumps, such as <i>Mar</i> operon or the MexAB-OprM resistance pump, had the same resistance to antibiotics when grown in biofilms as mutants that did not produce <i>Mar</i> or MexAB-OprM [[#References|[16]]]. <br> | |||
<br>Opposing research on gene expression in <i>P. aeruginosa</i> biofilms, however, shows that over ten genomic islands encode efflux pumps and are activated in the presence of antibiotics. This is interesting because it shows that multiple antibiotics, as well as drugs targeting the production and upkeep of biofilms, may be necessary to cure not only <i>P. aeruginosa</i> infections, but all chronic infections caused by bacterial (or joint bacterial and fungal infections) [[#References|[19]]]. <br> | |||
<br><b><u>Biofilms Provide Resistance </u></b><br> | |||
There are three main hypothesis on why antibiotics often do not successfully cure bacterial communities in biofilms. <br> | |||
<br><u>Failure to Diffuse</u><br> | |||
One method of resistance is due to the failure of antibiotics to fully diffuse through the biofilm. The slow the diffusion of antibiotics allows the antimicrobial agents to be deactivated in the outer layers of the biofilm. Even reactive oxidants, such as hydrogen peroxide, which are produced by phagocytic cells cannot diffuse through the entire biofilm, preventing biofilm death (Costerton, 1999). However, measurements of antibiotic diffusion in vitro have shown that a select few antibiotics readily fuse throughout the biofilm matrix, which is mostly water. If these antimicrobial agents are deactivated, however, the diffusion can be slowed down dramatically [[#References|[16]]]. <br> | |||
<br><u> Spatial Heterogeneity </u><br> | |||
Many of the cells exist in different metabolic states, meaning that at least one of the cells is guaranteed to survive a metabolically directed attack [[#References|[16]]]. <br> | |||
<br><i>Metabolic Inactive Zones</i><br> | |||
For example, some of the cells in the biofilm lack access to sufficient nutrients and therefore are slow growing; these slow growing bacteria are not extremely susceptible to microbial agents [[#References|[4]]]. pH differences can also occur due to accumulation of waste products. This could prevent the action of an antibiotic. This accumulation of acidic waste products can often cause bacterial cells to enter a non-growing state. Antibiotics, like Penicillin, which target cell wall synthesis, only work on growing bacteria. So, the accumulation of waste acid effectively stops the antibiotic from killing all of the bacterial in the biofilm, a necessity for recovery from pathogens. This hypothesis is supported by visualization of metabolically inactive zones within biofilms [[#References|[16]]]. <br> | |||
<br><i> Anaerobic niches</i><br> | |||
Anther example is that Oxygen is often completely consumed by the surface layers of a biofilm, creating anaerobic niches in the less exposed regions of the biofilm. These anaerobic niches are often unaffected by Aminoglycoside antibiotics [[#References|[16]]]. <br> | |||
<br><i>Osmotic Stress Response</i><br> | |||
<br> | Finally, the osmotic environment could be changed, causing an osmotic stress response. This response could change the amount of porins and cell envelope permeability, effectively stopping high numbers of antibiotics from gaining entrance [[#References|[16]]]. <br> | ||
<br><u>Distinct Resistive Phenotype</u><br> | |||
Thirdly, it is believed that some of the cells in biofilms adopt a distinct phenotype that protects them from antimicrobial agents. This phenotype, however, is not due to starvation, but is programmed based on environmental factors [[#References|[4]]]. This hypothesis is supported by studies that show resistance in new biofilms that are too thin to pose a physical barrier to antibiotic entrance. <br> | |||
<br>A major point is that all members of a biofilm must be removed or killed to prevent relapse, much in the same way that all cancer cells in a tumor must be removed or killed to prevent cancer reoccurrence. If as little as 1% of the original population persists after antimicrobial treatments, the infection can recur [[#References|[16]]]. <br> | |||
==Treatments== | |||
<b><u>Current therapies</u></b><br> | |||
<br><u>Antibiotics</u><br> | |||
Current therapies include strong courses of antibiotics, such as ciprofloxacin, which works by inhibiting DNA-gyrase in the bacterial cell. Additionally, consistent treatment with antibiotics to prevent colonization in young CF patients is promising | |||
[[#References|[4]]]. Macrolide antibiotics, such as azithromycin, have been shown to have anti-biofilm properties, although resistant mutants are readily selected | |||
[[#References|[23]]]<br> | |||
<u>Surgery</u><br> | |||
Antibiotics often fail to kill the bacterium in the biofilm; only planktonic bacteria that have detached and dispersed from the original biofilm, can be killed by antibiotics. For this reason, after antibiotic therapy is complete, the biofilm continues to produce planktonic bacteria and the infection reoccurs. For this reason, the only effective course of action is to surgically remove the biofilm if possibl | |||
[[#References|[4]]] <br> | |||
<u>Immunosuppression Drugs</u><br> | |||
Tissues near the biofilm may often become damaged by the excessive amount of neutrophils and immune complexes released in a healthy person’s immune response. This is the same reason the lungs of CF patients undergo much damage; the high number of antibodies produced often results in worsening health, as they cause inflammation and damage to pulmonary tissue | |||
[[#References|[16]]]. For this reason, immunosuppression drugs are often given to CF patients— the innate immune system cannot fight off the biofilms and doing so just worsens health conditions. However, consistent treatment with antibiotics to prevent colonization in young patients is promising | |||
[[#References|[4]]]. <br> | |||
<br><b><u>Potential Therapies</u></b><br> | |||
Due to the hetergenousity of biofilms, as well as there being multiple antibiotic resistance methods, it unlikely that any one antimicrobial agent would be able to kill all of the cells in the biofilm | |||
[[#References|[16]]] | |||
.<br> | |||
<u>Quorum Sensing Inhibitors</u><br> | |||
Quorum sensing is extremely important for the production and upkeep of biofilms. Because of this, the chemicals used in quorum sensing are excellent targets for new therapies. One way quorum sensing could be halted is to identify the chemicals responsible for the destruction of biofilms. The <i>BqsS-BqsR</i> system, identified by Yi-Hu Dong and Colleagues, does this. These genes regulate biofilm decay; if either of them are deleted or mutated, the rate of biofilm formation increased. This is because <i>BqsR</i> and <i>BqsS</i> are responsible for the production of certain Quorum sensing molecules, such as butyryl-L-homoserine lactone (C4HSL) and Pseudomonas quinolone signal (PQS), which reduce biofilm formation. When these chemicals are added to a biofilm, the rate of formation decreased [[#References|[8]]]. Therefore, if these molecules, or synthetic molecules similar in composition could be produced pharmaceutically, biofilms could be destroyed. No longer in a protected state, the immune system, or the immune system supplemented with antibiotics, could cure the infection.<br> | |||
<br> | <br> | ||
Another way quorum sensing could be halted is to identify the receptors and molecules important for the growth of biofilms. For example, two quorum sensing receptors, LasR and RhIR, are inhibited by meta-bromo-thiolactone (mBLT) in an experiment by O’Loughlin and colleagues. This compound inhibited the production of virulence factor pyocyanin, biofilm formation, as well as quorum sensing mediated death of host epithelial cells. [[#References|[12]]]. This is exciting because it, too, provides a new target for therapeutics. It is important to note that there has been little luck in blocking the aGM1 receptors <i>P. aeruginosa</i> use to adhere to host cells with; blocking thus receptor cause a significant inflammation response [[#References|[5]]] | |||
.<br> | |||
<br><u> Salt Resistant Antibiotics</u><br> | |||
Based on the hypothesis that the high amount of salt on the CF epithelial cells causes reduced immune response ability, antimicrobial agents built specifically to withstand high-salt conditions could be synthetically created or found in the environments | |||
[[#References|[5]]]. | |||
==References== | ==References== | ||
[[#References|[5]]] | |||
[1]Banin, E. Proceedings of the National Academy of Sciences - PNAS: Iron and Pseudomonas Aeruginosa Biofilm Formation. 102 Vol. The Academy, 08/02/2005. Web. 20 Apr. 2015. | |||
[2]Bals, R. The Journal of Clinical Investigation: Human Beta-Defensin 2 is a Salt-Sensitive Peptide Antibiotic Expressed in Human Lung. 102 Vol. American Society for Clinical Investigation, 09/01/1998. Web. 7 May 2015. | |||
[3] Bjarnsholt, Thomas. Pediatric Pulmonology: Biofilms in the Respiratory Tract of Cystic Fibrosis Patients. 44 Vol. John Wiley & Sons Inc, 06/2009. Web. 22 Apr. 2015. | |||
[4] Costerton, J. W. Science (New York, N.Y.): Bacterial Biofilms: A Common Cause of Persistent Infections. 284 Vol. American Association for the Advancement of Science, 05/21/1999. Web. 20 Apr. 2015. | |||
[5]Davies, Jane C. Paediatric Respiratory Reviews: Pseudomonas Aeruginosa in Cystic Fibrosis: Pathogenesis and Persistence. 3 Vol. Elsevier, 06/2002. Web. 7 May 2015. | |||
[6]Delgado, M. A., J. F. Poschet, and V. Deretic. "Nonclassical Pathway of Pseudomonas Aeruginosa DNA-Induced Interleukin-8 Secretion in Cystic Fibrosis Airway Epithelial Cells." Infection and Immunity 74.5 (2006): 2975-984. Web. | |||
[7]Donaldson, Scott H. The New England Journal of Medicine: Mucus Clearance and Lung Function in Cystic Fibrosis with Hypertonic Saline. 354 Vol. Massachusetts Medical Society, 01/2006. Web. 7 May 2015. | |||
[8]Dong, Yihu. Communicative & Integrative Biology: A Novel Two-Component System BqsS-BqsR Modulates Quorum Sensing-Dependent Biofilm Decay in. 1 Vol. Landes Bioscience, 07/2008. Web. 7 May 2015. | |||
[9]Harrison-Balestra, Catherine. Dermatologic Surgery: A Wound-Isolated Pseudomonas Aeruginosa Grows a Biofilm in Vitro within 10 Hours and is Visualized by Light Microscopy. 29 Vol. Blackwell Publishing, 06/2003. Web. 7 May 2015. | |||
[10] Lederberg, Joshua et al. Pseudomonas. Encyclopedia of Microbiology. Second Edition. Volume 3. San Diego, 2000. p. 876-891. | |||
[11] Oliver, A. Science (New York, N.Y.): High Frequency of Hypermutable Pseudomonas Aeruginosa in Cystic Fibrosis Lung Infection. 288 Vol. American Association for the Advancement of Science, 05/19/2000. Web. 22 Apr. 2015. | |||
[12]O'Loughlin, C. T. Proceedings of the National Academy of Sciences - PNAS: A Quorum-Sensing Inhibitor Blocks Pseudomonas Aeruginosa Virulence and Biofilm Formation. 110 Vol. The Academy, 10/29/2013. Web. 7 May 2015. | |||
[13]Rogers, C. S. Science (New York, N.Y.): Disruption of the CFTR Gene Produces a Model of Cystic Fibrosis in Newborn Pigs. 321 Vol. American Association for the Advancement of Science, 09/26/2008. Web. 7 May 2015. | |||
[14]Saiman, L., and A. Prince. "Pseudomonas Aeruginosa Pili Bind to AsialoGM1 Which Is Increased on the Surface of Cystic Fibrosis Epithelial Cells." Journal of Clinical Investigation 92.4 (1993): 1875-880. Web. | |||
[15]Smith, Jeffrey J. Cell (Cambridge): Cystic Fibrosis Airway Epithelia Fail to Kill Bacteria because of Abnormal Airway Surface Fluid. 85 Vol. Cell Press, 04/1996. Web. 7 May 2015. | |||
[16] Stewart, Philip S. The Lancet (North American Edition): Antibiotic Resistance of Bacteria in Biofilms. 358 Vol. Little, Brown and Co, 07/2001. Web. 20 Apr. 2015. | |||
[17]Trafny, Elżbieta Anna. International Journal of Antimicrobial Agents: Susceptibility of Adherent Organisms from Pseudomonas Aeruginosa and Staphylococcus Aureus Strains Isolated from Burn Wounds to Antimicrobial Agents. 10 Vol. Elsevier, 08/1998. Web. 7 May 2015. | |||
[18]"What Is Pseudomonas Aeruginosa?" Pseudomonas Aeruginosa. EHA Consulting Group, 2015. Web. 07 May 2015. | |||
[19] Whiteley, Marvin. Nature (London): Gene Expression in Pseudomonas Aeruginosa Biofilms. 413 Vol. Macmillan Journals Ltd., etc, 10/2001. Web. 20 Apr. 2015. | |||
[ | [20]Lessnau, Klaus-Dieter, and Michael MM Bronze. "Pseudomonas Aeruginosa Infections Clinical Presentation." Pseudomonas Aeruginosa Infections Clinical Presentation. Medscape, 28 Apr. 2014. Web. 07 May 2015. | ||
[ | [21]Johnson, J. Kristie. "The Role of Patient-to-Patient Transmission in the Acquisition of Imipenem-Resistant Pseudomonas Aeruginosa Colonization in the Intensive Care Unit." The Journal of Infectious Diseases 200.6 (2009): 900-05. JSTOR. Web. 07 May 2015. | ||
[ | [22]McCulloch, Elaine. Journal of Cystic Fibrosis: Improved Early Diagnosis of Pseudomonas Aeruginosa by Real-Time PCR to Prevent Chronic Colonisation in a Paediatric Cystic Fibrosis Population. 10 Vol. Elsevier, 01/2011. Web. 7 May 2015. | ||
[ | [23]Mulet, X. Antimicrobial Agents and Chemotherapy: Azithromycin in Pseudomonas Aeruginosa Biofilms: Bactericidal Activity and Selection of nfxB Mutants. 53 Vol. American Society for Microbiology, 04/2009. Web. 7 May 2015. | ||
<br><br>Authored for [http://biology.kenyon.edu/courses/biol238/biol238syl15.html BIOL 238 Microbiology], taught by [mailto:slonczewski@kenyon.edu Joan Slonczewski], 2015, [http://www.kenyon.edu/index.xml Kenyon College]. | <br><br>Authored for [http://biology.kenyon.edu/courses/biol238/biol238syl15.html BIOL 238 Microbiology], taught by [mailto:slonczewski@kenyon.edu Joan Slonczewski], 2015, [http://www.kenyon.edu/index.xml Kenyon College]. | ||
Latest revision as of 20:26, 29 September 2015
Introduction
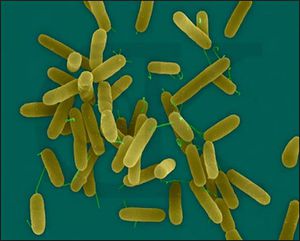
P. aeruginosa is a gram negative bacterium with one flagella, as can be seen in Figure 1 [10]. The flagellum plays an extremely important role in the production and assembly of biofilm formation. P. aeruginosa is also a facultative anaerobe, but its optimal metabolism is aerobic respiration. In the absence of oxygen, it can respire anaerobically using nitrate or an alternate electron acceptor [4]. However, it often cannot grow when it is simultaneously a pathogen and not exposed to oxygen, as is common in the depth of the biofilm. Many cells thus often remain in an un-dividing state when in a biofilm. This, however, works in P. aeruginosa’s favor, as many antibiotics cannot kill cells that are not actively growing or dividing [16]. Furthermore, it can catabolize a variety of organic molecules, even carcinogens such as benzoate. This incredible nutritional and metabolic versatility and diversity allows P. aeruginosa to survive attack by both phage and human induced antimicrobial agents, as well as live in a diverse array of environments. P. aeruginosa is found ubiquitously, in environments as diverse as hospitals, sewage, animals, plants, water, and the soil. It is also the most predominant organism in oligotrophic aquatic ecosystems. Furthermore, in the soil, P. aeruginosa is an important ecological bacterial species, breaking down the polycyclic aromatic hydrocarbons, and catabolizing important molecules such as rhamnolipids, quinolones, and hydrogen cyanide [10].This versatility allows it to be the most abundant organism on earth.
Despite this abundance, it is an opportunistic pathogen, rarely infecting healthy individuals. It is common in immunocompromised patients, such as people suffering from Acquired Immunodeficiency Syndrome (AIDS) or cancer, as well as burn victim (Davis, 2003; Lederberg, 2000). Additionally, many Cystic Fibrosis (CF) patients are chronically infected by P. aeruginosa, causing severe pulmonary inflammation and damage. There are many hypotheses explaining the abundance of P. aeruginosa infections in the CF population, but the most well supported is the high-salt hypotheis, which states that high rate of infection are due to CF patients having an abundance of salt in their respiratory tract; this prevents the production and action of chemicals and proteins used by the immune system to prevent infection [5] [16]. Because of its prevalence in hospitals and selective pathogenesis, P. aeruginosa infects two thirds of critically ill hospitalized patients.
Unfortunately, it is an extremely invasive pathogen, with a mortality rate of 40-60%. The mortality rate is extremely high because of the biofilm nature of P. aeruginosa. When a pathogen is present in the human lungs, it often forms biofilms, communities of bacterial cells attached to an inert or living surface and enclosed in an extracellular polysaccharide matrixes. These biofilms release planktonic bacteria, which disperse to spread the infection to other tissues or host organisms. Many antibiotics kill the planktonic bacteria. However, antibiotics fail to kill the bacterium in the biofilm; for this reason, after antibiotic therapy is complete, the biofilm continues to produce planktonic bacteria and the infection reoccurs. Biofilms are inherently resistant to antibiotics, whether in the form of bacteriophages, amoebae, or chemical biocides [10].
Pathology
Clinical Feature
P. Aeruginosa can infect may parts of the body. It is most often seen, however, in the respiratory tract due to its prevalence in the lower respiratory tract of the CF population. Here it generally causes pneumonia, which has symptoms such as fever, cyanosis, the chills, and a productive cough. It is however also seen in the blood, which is usually acquired from medical devices. It may also be acquired from native and prosthetic heart valves, causing endocarditis. Causing meningitis and brain abscesses, it also infects the Central Nervous System. This too, is usually the result of a medical procedure, such as paranasal sinus surgery. When infecting the eye, P. aeruginosa, produces extracellular enzymes that cause rapid destruction of the infected lesion. This causes many forms of vision impairment, such as bacterial keratitis, scleral abscesses, and ophthalmic neonatorum. It can also be found in the ear, skin, gastrointestinal tract, urinary tract, and bones [20].
Transmission and Diagnosis
P. aeruginosa is an ubiquitous organism, found in diverse environments, such as hospitals, sewage, animals, plants, water, and the soil (Lederberg, 2000). It is also the most predominant organism in oligotrophic aquatic ecosystems. Despite, this abundance, P. aeruginosa is an opportunistic pathogen, rarely infecting healthy individuals (Stewart, 2001). Infections occur in patients with compromised immune systems, such as people with acquired immune deficiency syndrome (AIDS). They also occur when the bacterium surpass the physical barriers of the innate immune system, which can be seen in the high rate of infections in burn victims [5]. In most cases, P. aeruginosa infections spread though contact with other, infected individuals. One study by Johnson and colleagues used gel electrophoresis to find that in a selected Intensive Care Unit, 31% of Imipenem resistant P. aeruginosa cases were due to patient-to-patient transmission. Even more interestingly, 19% of cases were from the patients’ own endogenous flora [21].
The diagnosis of P.aeruginosa infections can be made several ways based on its identifying features. The bacterium grows well on laboratory media, especially eosin-methylthionine blue agar and blood agar. It can be identified based on it Gram negative morphology, fruity odor, and ability to grow at high temperature, like 42 degrees C. Due to the chemical pyocyanin, it can florescence blue under ultraviolet light. This helps with its early identification in wounds [18]. Finally, real time Polymerase Chain Reaction (PCR) of cough and septum swabs is a new, exciting way to quickly and accurately detect the presence of P. aeruginosa [22].
Once infected with P. aeruginosa, the prognosis is poor: it has a mortality rate of 40-60%. This mortality rate is due to the biofilm nature of P. aeruginosa. When a pathogen colonizes human lungs, it often forms biofilms, communities of bacterial cells attached to an inert or living surface and enclosed in an extracellular polysaccharide matrixes. These biofilms release planktonic bacteria, which are dispersed to spread the infection to other tissues or host organism. Many antibiotics kill the planktonic bacteria. However, antibiotics fail to kill the bacterium in the biofilm; for this reason, after antibiotic therapy is complete, the biofilm continues to produce planktonic bacteria and the infection reoccurs [10]. This reoccurring infection results slowly destroys the infected organs through the release of toxins.
In Burn Victims
P. aeruginosa is the most common form of infection suffered by burn victims. Human skin is comprised of two layers, the epidermal and dermis. When the skin is burned, it allows microbes to colonize the subcutaneous tissue. In an experiment done by Trafny and colleagues, animals with partial-thickness cutaneous burns developed mature biofilms in 48 to 72 hours [17]. P.aeruginosa biofilms have been shown to be fully mature in humans only 10 hours after burns [9].
In Cystic Fibrosis (CF) patients
CF is an inherited autosomal recessive disease that results from mutations in the gene encoding the CF transmembrane conductance regulator (CFTR) protein.
CFTR is a bidirectional chloride channel and unidirectional sodium channel in epithelial cells [5]. If this protein is not fully functional, chloride cannot move into and out of the cells and sodium cannot cross the cell membrane; as chloride is important for water movement, mucus becomes thick.
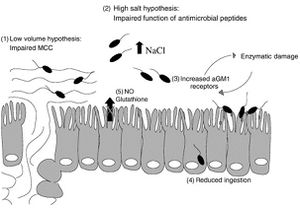
In healthy lungs, entering P. aeruginosa are systematically removed or killed quickly by a variety of innate immune responses, such as mucociliary clearance and macrophages. However, when CF patients contract P. aeruginosa, it causes chronic infection. 85% of CF patients have chronic [5]P. aeruginosa infections by adolescence. This elicits an aggressive inflammatory response, which causes lung damage. There are currently several hypotheses explaining the prevalence of P. aeruginosa, which are diagramed in Figure 3. [7]
Decreased Clearance of Mucociliary Passages
To begin, some propose that the sodium and water hyperabsorption in CF causes the airway surface liquid (ASL) and mucus layer to have an abnormally small volume. This causes the cilia to beat inefficiently, impairing the ability of CF patients to clear their mucociliary passages (throat). This causes inhaled pathogens to become trapped, promoting inflammation and infection [5]. Experiments have shown that treatment with hypertonic saline improves lung function and mucus clearance rate, which could reduce the occurrence of bacterial entrapment causing P. aeruginosa infection [7].
High Salt Deactivation Hypothesis
A second hypothesis centers on the high levels of sodium and chloride in the ASL. Many immune defenses, such as hBD-1 lysozymes and lactoferrin, are salt sensitive. Smith and colleagues showed that many of these defensives are rendered nonfunctional in high salt environments, like CF lungs, using an in vitro experiment [15]. Another recent experiment by Bals and colleagues showed that hBD-2, a peptide with antimicrobial properties expressed throughout the epithelium, is sensitive to salt concentrations; its ability to inhibit bacterial growth diminished as the concentration of NaCl increased. Its ability to function in salt was tested by exposing E. coli cells to isolated hBD-2 peptide at differing NaCl concentration [2]. This suggests that it would be nonfunctional in CF and this breach in the innate immunity could be the cause of many infections.
Acidic Environment Hypothesis
Another possibility is that the acidic pH of CF epithelium impairs mechanisms of the immune systems. CFTR is involved in the transport of bicarbonate ions, which regulate cell surface pH, making CF epithelium abnormally acidic. This may stop immune system mechanisms, such as the mucociliary clearance or phagocyte function from fully working [5]. While the decrease in ASL pH has not been shown conclusively in humans, Pezzulo and colleagues discovered that pigs lacking CFTR have more acidic ASL compared to pigs with functional CFTR [13]. Furthermore, IL-8 is secreted in response to P. aeruginosa and causes severe inflammation. Its secretion, however, decreased with treatments of substances that inhibit acidification of intracellular organelles such as chloroquine. This shows that either P. aeruginosa is less effective in basic conditions, which is unlikely considering the range of environments it resides in, or that the immune system is more inept at less acidic conditions [6].
aGMI Receptor Hypothesis
P. aeruginosa used several different types of organelles, including pili and flagella for adherance to cell surface receptors. The receptor for both pili and flagella is GalNAcβ1-4Gal, which consists of several glycolipids. Studies, such as one done by Saiman and colleagues, has shown that one of the glycoproteins, asialoGM1 (aGM1), is present in a higher concentration on CF epithelial cells compared to normal cells (12% v 2.9% , respectively) [14].
Inflammation Hypothesis
Finally, research has shown that when P. aeruginosa DNA is released from the bacterium, it causes inflammation of CF epithelial cells. It does this by using p38 and Erk mitogen-activated protein kinase to induce the secretion of interleukin-8 (IL-8) from the host cells. This chemokine causes excessive neutrophil infiltration. This was determined in vitro by exposing bronchial epithelial cell lines with and without functional CTFR to P. aeruginosa and then quantifying the gene and protein products secreted [6]. This research indicates that P. aeruginosa could cause inflammation of the epithelial cells without the bacteria even adhering to the cell surface [5].
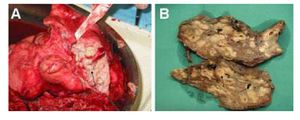
Tissues near the infected biofilm often become severely damages in CF patients due to the excessive amount of neutrophils and immune complexes, such as IL-8, released, as well as the toxins produced by the P. aeruginos a bacterium [16]. This can be seen in Figure 2: the lungs are incredibly damaged. Because of the damage to pulmonary tissue, immunosuppression drugs are often given to CF patients— the innate immune system cannot fight off the biofilms and doing so just worsens health conditions. However, consistent treatment with antibiotics to prevent colonization in young patients is promising [4].
In Hospitals
Due to the high presence of immunocompromised patients, P. aeruginosa infections are very common in hospitals. It infects two thirds of critically ill hospitalized patients [10]. Much of the transmission is from biofilms that have colonized on medical devices. Pathogenic biofilms have been found using electron microscopy on the surface of medical devices, such as central venous catheters, prosthetic heart valves, and orthopedic implants (Costerton, 1999; Stewart, 2001). However, other studies, such as that done by Johnson and colleagues in a selected Intensive Care Unit showed that 31% of Imipenem resistant P. aeruginosa cases were due to patient-to-patient transmission. Even more interesting, 19% of cases were from the patients’ own endogenous flora [21].
Biofilms
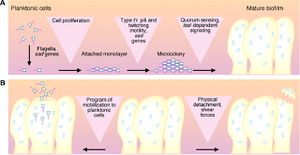
Biofilms are organized communities of bacterial cells attached to an inert or living surface and enclosed in an extracellular polysaccharide matrix. This matrix forms a slippery, solid coat around the bacterial community used to protect the bacterial community in a hostile environment, such as the human lungs, where pathogens are systematically removed by a variety of innate immune responses, such as mucociliary clearance [5]. These bacterial communities cause many persistent and chronic bacterial infections, such as P. aeruginosa infection in immunocompromised patients. An understanding of both biofilm production and interaction, therefore, is important because it could provide new therapeutic targets [4].
Life Cycle of P. aeruginosa
Bacterial Attachment
The initial attachment requires both flagella, as show by an experiment done by O’Toole and Kolter, where knock out mutants without flagella could not adequately adhere to a surface. During this attachment phase, the transcription of particular genes is activated, such as algC, algD, and algU, which are required for the synthesis of the extracellular polysaccharide that encases the biofilm. This is spectacular because it implies that P. aeruginosa have a ‘sense of touch’ that enables the detection and reaction to a surface [4].
Formation of Microcolonies
Next, microcolonies are formed. Microcolonies are relatively small groups of bacteria. The formation of microcolonies requires Type IV pili, as shown by O’Toole and Kolter. Bacteria lacking pili were able to form a monolayer, but not microcolonies. These pili are involved in a ‘twitching,’ which is thought to be necessary to the aggregation of cells into microcolonies [4].
Differentiation
These microcolonies then differentiate into true biofilms. Quorum sensing, the exchange of chemical signals between cells, may affect the timing of this differentiation as well as the coordinated building of the biofilm into complex structures [4].
Bloodstream Dissemination
Finally, the planktonic cells detach and differentiate from the biofilms. One mechanism for this to occur is that the pieces of biofilms break off, but this break may be regulated based on molecular cues; there appears to be a programmed detachment of planktonic bacteria. For example, it is thought that the release of planktonic cells requires an enzyme to digest alginate [4]. Figure 4 outlines this entire process.
Requirements for Biofilm Formation
Gene Expression
P. aeruginosa planktonic cells and biofilm cells have extremely different lifestyles. Planktonic cells are not surrounded by an extracellular polysaccharide matrix, like their biofilm enclosed counterparts, but instead dissociate and diffuse from the biofilm in order to spread the bacteria to new tissues and host organisms. Because of this, it is surprising that the two types of P. aeruginosa cells differ only by 1% in terms of differential gene expression. This refutes the argument that P. aeruginosa’s existence in a biofilm is dependent on dramatic differences in gene expression. However, the small amount of genes that are activated and repressed (34 and 39 respectively) may be crucial for the aggregation of the cells into a biofilm.
Gene specifying pili and flagella are suppressed in biofilm bacteria, suggesting that they are not needed for the incorporation and upkeep of a mature biofilm. They are instead probably important only in the initial process of biofilm development. Furthermore, the genes with the highest expression in the biofilm P. aeruginosa were those from a bacteriophage related to Pf1. Pf1 genes were activated 100 to 1000 times more in biofilms than planktonic bacteria. This shows that phage induction is important for gene transfer within biofilms. These genes are suspected to function either to exclude certain strains of P. aeruginosa from biofilms induction or to contain a gene specifying an important toxin [19].
Quorum Sensing
Quorum sensing is thought to determine when P. aeruginosa and other Gram Negative microcolonies differentiate into true biofilms. Individual cells produce acylhomoserine lactone signals; when a sufficient number of acylhomoserine producing cells are present, the signals accumulate and trigger the transcription and translation of a specific set of genes (Costerton, 1999).
LasR-LasI is a gene that is also related to quorum sensing and differentiation in P. aeruginosa. It controls the expression of extracellular virulence factors, as well as the Rh1R and Rh1I, which control the expression of many secondary metabolites, such as butyrylhomoserine. Mutants of this process do not differentiate [4].
It is believed that if cell-to-cell signaling is blocked or destroyed, biofilm production and maintenance would cease. If the biofilm were destroyed, it could allow the host immune system (or the host immune system supplemented with traditional antibiotics) to destroy the P. aeruginosa cells, in much the same way that antibiotics kill planktonic P. aeruginosa cells. [5][16].
Iron and Biofilm Production
Iron is a signal for biofilm production in P. aeruginosa. It is believed that either the level of internal iron or the active transport of chelated iron acts as a signal to produce biofilm formation. Iron starvation can often prevent the growth of bacteria, but sufficient levels may serve as a signal for biofilm development. This has been demonstrated by Lactoferrin, which sequesters iron and prevents P. aeruginosa from maturing from thin monolayers into multicellular biofilm structures. This could be due to the fact that low iron causes continuous twitching movements, the control of which is needed to form biofilm structures. Iron is often acquired by pyverdine, which is regulated by PvdS. When pyverdine binds to an outer-membrane receptor, it results in a signal that governs not only the activity of PvdS, but extracellular virulence factors. This is important for the opportunistic nature of P. aeruginosa infections; only when iron concentration is high and the cell is tethered to a surface, can pathogenic biofilm production begin. The Fur protein also regulates iron-responsive genes through a pair of small regulatory RNAs. The authors found that any pathway that allows iron into the cell –pryverdine or Fur, for example— can function as an iron-signaling pathway (and thus as a signal for virulence and biofilm development pathway). This indicates that a critical content of intracellular iron is a signal for biofilm pathway initiation [1].
Antibiotic Resistance
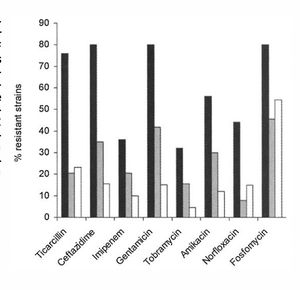
Biofilms are inherently resistant to antibiotics, whether in the form of bacteriophages, amoebae, or chemical biocides. They have this resistance through multiple mechanisms; however, some antibiotics are more effective than others at penetrating and killing cells in the biofilm (figure 3) [16].
Efflux Pumps
Often, bacteria avoid antibiotic-caused bactericide through the use of efflux pumps. However, these mutations are not the sole cause of antimicrobial protection in P. aeruginosa biofilms.
Studies on genes encoding efflux pumps, for example, revealed that P. aeruginosa with multidrug efflux pumps, such as Mar operon or the MexAB-OprM resistance pump, had the same resistance to antibiotics when grown in biofilms as mutants that did not produce Mar or MexAB-OprM [16].
Opposing research on gene expression in P. aeruginosa biofilms, however, shows that over ten genomic islands encode efflux pumps and are activated in the presence of antibiotics. This is interesting because it shows that multiple antibiotics, as well as drugs targeting the production and upkeep of biofilms, may be necessary to cure not only P. aeruginosa infections, but all chronic infections caused by bacterial (or joint bacterial and fungal infections) [19].
Biofilms Provide Resistance
There are three main hypothesis on why antibiotics often do not successfully cure bacterial communities in biofilms.
Failure to Diffuse
One method of resistance is due to the failure of antibiotics to fully diffuse through the biofilm. The slow the diffusion of antibiotics allows the antimicrobial agents to be deactivated in the outer layers of the biofilm. Even reactive oxidants, such as hydrogen peroxide, which are produced by phagocytic cells cannot diffuse through the entire biofilm, preventing biofilm death (Costerton, 1999). However, measurements of antibiotic diffusion in vitro have shown that a select few antibiotics readily fuse throughout the biofilm matrix, which is mostly water. If these antimicrobial agents are deactivated, however, the diffusion can be slowed down dramatically [16].
Spatial Heterogeneity
Many of the cells exist in different metabolic states, meaning that at least one of the cells is guaranteed to survive a metabolically directed attack [16].
Metabolic Inactive Zones
For example, some of the cells in the biofilm lack access to sufficient nutrients and therefore are slow growing; these slow growing bacteria are not extremely susceptible to microbial agents [4]. pH differences can also occur due to accumulation of waste products. This could prevent the action of an antibiotic. This accumulation of acidic waste products can often cause bacterial cells to enter a non-growing state. Antibiotics, like Penicillin, which target cell wall synthesis, only work on growing bacteria. So, the accumulation of waste acid effectively stops the antibiotic from killing all of the bacterial in the biofilm, a necessity for recovery from pathogens. This hypothesis is supported by visualization of metabolically inactive zones within biofilms [16].
Anaerobic niches
Anther example is that Oxygen is often completely consumed by the surface layers of a biofilm, creating anaerobic niches in the less exposed regions of the biofilm. These anaerobic niches are often unaffected by Aminoglycoside antibiotics [16].
Osmotic Stress Response
Finally, the osmotic environment could be changed, causing an osmotic stress response. This response could change the amount of porins and cell envelope permeability, effectively stopping high numbers of antibiotics from gaining entrance [16].
Distinct Resistive Phenotype
Thirdly, it is believed that some of the cells in biofilms adopt a distinct phenotype that protects them from antimicrobial agents. This phenotype, however, is not due to starvation, but is programmed based on environmental factors [4]. This hypothesis is supported by studies that show resistance in new biofilms that are too thin to pose a physical barrier to antibiotic entrance.
A major point is that all members of a biofilm must be removed or killed to prevent relapse, much in the same way that all cancer cells in a tumor must be removed or killed to prevent cancer reoccurrence. If as little as 1% of the original population persists after antimicrobial treatments, the infection can recur [16].
Treatments
Current therapies
Antibiotics
Current therapies include strong courses of antibiotics, such as ciprofloxacin, which works by inhibiting DNA-gyrase in the bacterial cell. Additionally, consistent treatment with antibiotics to prevent colonization in young CF patients is promising
[4]. Macrolide antibiotics, such as azithromycin, have been shown to have anti-biofilm properties, although resistant mutants are readily selected
[23]
Surgery
Antibiotics often fail to kill the bacterium in the biofilm; only planktonic bacteria that have detached and dispersed from the original biofilm, can be killed by antibiotics. For this reason, after antibiotic therapy is complete, the biofilm continues to produce planktonic bacteria and the infection reoccurs. For this reason, the only effective course of action is to surgically remove the biofilm if possibl
[4]
Immunosuppression Drugs
Tissues near the biofilm may often become damaged by the excessive amount of neutrophils and immune complexes released in a healthy person’s immune response. This is the same reason the lungs of CF patients undergo much damage; the high number of antibodies produced often results in worsening health, as they cause inflammation and damage to pulmonary tissue
[16]. For this reason, immunosuppression drugs are often given to CF patients— the innate immune system cannot fight off the biofilms and doing so just worsens health conditions. However, consistent treatment with antibiotics to prevent colonization in young patients is promising
[4].
Potential Therapies
Due to the hetergenousity of biofilms, as well as there being multiple antibiotic resistance methods, it unlikely that any one antimicrobial agent would be able to kill all of the cells in the biofilm
[16]
.
Quorum Sensing Inhibitors
Quorum sensing is extremely important for the production and upkeep of biofilms. Because of this, the chemicals used in quorum sensing are excellent targets for new therapies. One way quorum sensing could be halted is to identify the chemicals responsible for the destruction of biofilms. The BqsS-BqsR system, identified by Yi-Hu Dong and Colleagues, does this. These genes regulate biofilm decay; if either of them are deleted or mutated, the rate of biofilm formation increased. This is because BqsR and BqsS are responsible for the production of certain Quorum sensing molecules, such as butyryl-L-homoserine lactone (C4HSL) and Pseudomonas quinolone signal (PQS), which reduce biofilm formation. When these chemicals are added to a biofilm, the rate of formation decreased [8]. Therefore, if these molecules, or synthetic molecules similar in composition could be produced pharmaceutically, biofilms could be destroyed. No longer in a protected state, the immune system, or the immune system supplemented with antibiotics, could cure the infection.
Another way quorum sensing could be halted is to identify the receptors and molecules important for the growth of biofilms. For example, two quorum sensing receptors, LasR and RhIR, are inhibited by meta-bromo-thiolactone (mBLT) in an experiment by O’Loughlin and colleagues. This compound inhibited the production of virulence factor pyocyanin, biofilm formation, as well as quorum sensing mediated death of host epithelial cells. [12]. This is exciting because it, too, provides a new target for therapeutics. It is important to note that there has been little luck in blocking the aGM1 receptors P. aeruginosa use to adhere to host cells with; blocking thus receptor cause a significant inflammation response [5]
.
Salt Resistant Antibiotics
Based on the hypothesis that the high amount of salt on the CF epithelial cells causes reduced immune response ability, antimicrobial agents built specifically to withstand high-salt conditions could be synthetically created or found in the environments
[5].
References
[5] [1]Banin, E. Proceedings of the National Academy of Sciences - PNAS: Iron and Pseudomonas Aeruginosa Biofilm Formation. 102 Vol. The Academy, 08/02/2005. Web. 20 Apr. 2015.
[2]Bals, R. The Journal of Clinical Investigation: Human Beta-Defensin 2 is a Salt-Sensitive Peptide Antibiotic Expressed in Human Lung. 102 Vol. American Society for Clinical Investigation, 09/01/1998. Web. 7 May 2015.
[3] Bjarnsholt, Thomas. Pediatric Pulmonology: Biofilms in the Respiratory Tract of Cystic Fibrosis Patients. 44 Vol. John Wiley & Sons Inc, 06/2009. Web. 22 Apr. 2015.
[4] Costerton, J. W. Science (New York, N.Y.): Bacterial Biofilms: A Common Cause of Persistent Infections. 284 Vol. American Association for the Advancement of Science, 05/21/1999. Web. 20 Apr. 2015.
[5]Davies, Jane C. Paediatric Respiratory Reviews: Pseudomonas Aeruginosa in Cystic Fibrosis: Pathogenesis and Persistence. 3 Vol. Elsevier, 06/2002. Web. 7 May 2015.
[6]Delgado, M. A., J. F. Poschet, and V. Deretic. "Nonclassical Pathway of Pseudomonas Aeruginosa DNA-Induced Interleukin-8 Secretion in Cystic Fibrosis Airway Epithelial Cells." Infection and Immunity 74.5 (2006): 2975-984. Web.
[7]Donaldson, Scott H. The New England Journal of Medicine: Mucus Clearance and Lung Function in Cystic Fibrosis with Hypertonic Saline. 354 Vol. Massachusetts Medical Society, 01/2006. Web. 7 May 2015.
[8]Dong, Yihu. Communicative & Integrative Biology: A Novel Two-Component System BqsS-BqsR Modulates Quorum Sensing-Dependent Biofilm Decay in. 1 Vol. Landes Bioscience, 07/2008. Web. 7 May 2015.
[9]Harrison-Balestra, Catherine. Dermatologic Surgery: A Wound-Isolated Pseudomonas Aeruginosa Grows a Biofilm in Vitro within 10 Hours and is Visualized by Light Microscopy. 29 Vol. Blackwell Publishing, 06/2003. Web. 7 May 2015.
[10] Lederberg, Joshua et al. Pseudomonas. Encyclopedia of Microbiology. Second Edition. Volume 3. San Diego, 2000. p. 876-891.
[11] Oliver, A. Science (New York, N.Y.): High Frequency of Hypermutable Pseudomonas Aeruginosa in Cystic Fibrosis Lung Infection. 288 Vol. American Association for the Advancement of Science, 05/19/2000. Web. 22 Apr. 2015.
[12]O'Loughlin, C. T. Proceedings of the National Academy of Sciences - PNAS: A Quorum-Sensing Inhibitor Blocks Pseudomonas Aeruginosa Virulence and Biofilm Formation. 110 Vol. The Academy, 10/29/2013. Web. 7 May 2015.
[13]Rogers, C. S. Science (New York, N.Y.): Disruption of the CFTR Gene Produces a Model of Cystic Fibrosis in Newborn Pigs. 321 Vol. American Association for the Advancement of Science, 09/26/2008. Web. 7 May 2015.
[14]Saiman, L., and A. Prince. "Pseudomonas Aeruginosa Pili Bind to AsialoGM1 Which Is Increased on the Surface of Cystic Fibrosis Epithelial Cells." Journal of Clinical Investigation 92.4 (1993): 1875-880. Web.
[15]Smith, Jeffrey J. Cell (Cambridge): Cystic Fibrosis Airway Epithelia Fail to Kill Bacteria because of Abnormal Airway Surface Fluid. 85 Vol. Cell Press, 04/1996. Web. 7 May 2015.
[16] Stewart, Philip S. The Lancet (North American Edition): Antibiotic Resistance of Bacteria in Biofilms. 358 Vol. Little, Brown and Co, 07/2001. Web. 20 Apr. 2015.
[17]Trafny, Elżbieta Anna. International Journal of Antimicrobial Agents: Susceptibility of Adherent Organisms from Pseudomonas Aeruginosa and Staphylococcus Aureus Strains Isolated from Burn Wounds to Antimicrobial Agents. 10 Vol. Elsevier, 08/1998. Web. 7 May 2015.
[18]"What Is Pseudomonas Aeruginosa?" Pseudomonas Aeruginosa. EHA Consulting Group, 2015. Web. 07 May 2015.
[19] Whiteley, Marvin. Nature (London): Gene Expression in Pseudomonas Aeruginosa Biofilms. 413 Vol. Macmillan Journals Ltd., etc, 10/2001. Web. 20 Apr. 2015.
[20]Lessnau, Klaus-Dieter, and Michael MM Bronze. "Pseudomonas Aeruginosa Infections Clinical Presentation." Pseudomonas Aeruginosa Infections Clinical Presentation. Medscape, 28 Apr. 2014. Web. 07 May 2015.
[21]Johnson, J. Kristie. "The Role of Patient-to-Patient Transmission in the Acquisition of Imipenem-Resistant Pseudomonas Aeruginosa Colonization in the Intensive Care Unit." The Journal of Infectious Diseases 200.6 (2009): 900-05. JSTOR. Web. 07 May 2015.
[22]McCulloch, Elaine. Journal of Cystic Fibrosis: Improved Early Diagnosis of Pseudomonas Aeruginosa by Real-Time PCR to Prevent Chronic Colonisation in a Paediatric Cystic Fibrosis Population. 10 Vol. Elsevier, 01/2011. Web. 7 May 2015.
[23]Mulet, X. Antimicrobial Agents and Chemotherapy: Azithromycin in Pseudomonas Aeruginosa Biofilms: Bactericidal Activity and Selection of nfxB Mutants. 53 Vol. American Society for Microbiology, 04/2009. Web. 7 May 2015.
Authored for BIOL 238 Microbiology, taught by Joan Slonczewski, 2015, Kenyon College.
