Pseudomonas Aeruginosa Infection and Biofilm Production in Immunocompromised Individuals: Difference between revisions
| Line 32: | Line 32: | ||
<br> | <br> | ||
==Antibiotic Resistance== | ==Antibiotic Resistance== | ||
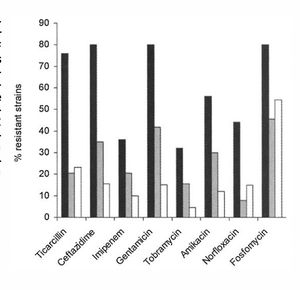
[[Image:anti.jpg|thumb|300px|right| This figure shows the differeences in antibiotic resistance between mutator CF cells (black), nonmutator CF cells (grey) and non-CF cells (white). There are significant differences between the mutator and non-mutator CF cells when immersed in ticarcillin, ceftazidime, gentamicin, amikacin, norfloxacin, and fosfomycin, all antibiotics. Oliver, A. Science (New York, N.Y.): High Frequency of Hypermutable Pseudomonas Aeruginosa in Cystic Fibrosis Lung Infection. 288 Vol. American Association for the Advancement of Science, 05/19/2000. Web. 22 Apr. 2015.]] | |||
Often, bacteria avoid bactericide by antibiotic using efflux pumps, modifying enzymes, or targeted mutations. However, these mutations are not what cause antimicrobial protection in biofilms. Studies on genes encoding efflux pumps, for example, revealed that P. aeruginosa with multidrug efflux pumps, such as Mar operon or the MexAB-OprM resistance pump, had the same resistance to antibiotics when grown in biofilms as mutant that did not produce Mar or MexAB-OprM (Stewart, 2001). Opposing research on gene expression in biofilms, however, dhows that over ten genomic islands encode efflux pumps and are activated in the presence of antibiotics. This is interesting because it shows that multiple antibiotics, as well as drugs targeting the production and upkeep of biofilms, may be necessary to cure not only P. aeruginosa infections, but all chronic infections caused by bacterial (or joint bacterial and fungal infections) (Whitely, 2001). Many antibiotics kill the planktonic bacteria released, but fail to kill the bacterium in the biofilm; for this reason, after antibiotic therapy is complete, the biofilm continues to produce planktonic bacteria and the infection reoccurs. There even appears to be a programmed detachment of planktonic bacteria. Biofilms are inherently resistant to antibiotics, weather in the form of bacteriophages, amoebae, or chemical biocides. They have this resistance through multiple mechanisms. The fact that the planktonic bacteria are killed by antibiotics suggests that resistance of biofilms is not due to mobile genetic elements mutations (Costerton, 1999).<br> | Often, bacteria avoid bactericide by antibiotic using efflux pumps, modifying enzymes, or targeted mutations. However, these mutations are not what cause antimicrobial protection in biofilms. Studies on genes encoding efflux pumps, for example, revealed that P. aeruginosa with multidrug efflux pumps, such as Mar operon or the MexAB-OprM resistance pump, had the same resistance to antibiotics when grown in biofilms as mutant that did not produce Mar or MexAB-OprM (Stewart, 2001). Opposing research on gene expression in biofilms, however, dhows that over ten genomic islands encode efflux pumps and are activated in the presence of antibiotics. This is interesting because it shows that multiple antibiotics, as well as drugs targeting the production and upkeep of biofilms, may be necessary to cure not only P. aeruginosa infections, but all chronic infections caused by bacterial (or joint bacterial and fungal infections) (Whitely, 2001). Many antibiotics kill the planktonic bacteria released, but fail to kill the bacterium in the biofilm; for this reason, after antibiotic therapy is complete, the biofilm continues to produce planktonic bacteria and the infection reoccurs. There even appears to be a programmed detachment of planktonic bacteria. Biofilms are inherently resistant to antibiotics, weather in the form of bacteriophages, amoebae, or chemical biocides. They have this resistance through multiple mechanisms. The fact that the planktonic bacteria are killed by antibiotics suggests that resistance of biofilms is not due to mobile genetic elements mutations (Costerton, 1999).<br> | ||
<br> One method of resistance is due to the failure of the antibiotic to fully diffuse through the biofilm. The slow the diffusion of antibiotics, allowing the antimicrobial agents to be deactivate in the outer layers of the biofilm. Even reactive oxidants, such as hydrogen peroxide, which are produced by phagocytic cells cannot diffuse through the entire biofilm, preventing biofilm death (Costerton, 1999). However, measurements of antibiotic diffusion in vitro have shown that a select few antibiotics readily fuse throughout the biofilm matrix, which is mostly water. If these antimicrobial agents are deactivator, however the diffusion can be slowed down dramatically (Stewart, 2001). | <br> One method of resistance is due to the failure of the antibiotic to fully diffuse through the biofilm. The slow the diffusion of antibiotics, allowing the antimicrobial agents to be deactivate in the outer layers of the biofilm. Even reactive oxidants, such as hydrogen peroxide, which are produced by phagocytic cells cannot diffuse through the entire biofilm, preventing biofilm death (Costerton, 1999). However, measurements of antibiotic diffusion in vitro have shown that a select few antibiotics readily fuse throughout the biofilm matrix, which is mostly water. If these antimicrobial agents are deactivator, however the diffusion can be slowed down dramatically (Stewart, 2001). | ||
| Line 37: | Line 39: | ||
<br> Thirdly, it is believed that some of the cells in biofilms adopt a distinct phenotype that protects them from antimicrobial agents. This phenotype, however, is not due to starvation, but is programmed based on environmental factors (Costerton, 1999). This hypothesis is supported by studies that show resistance in new biofilms that are too thin to pose a physical barrier to antibiotic entrance. A major point is that all members of a biofilm must be removed or killed to prevent relapse, much in the same way that all cancer cells in a tumor must be removed or killed to prevent cancer reoccurrence. If as little as 1% of the original population persists after antimicrobial treatments, the infection can come back (Stewart, 2001). <br> | <br> Thirdly, it is believed that some of the cells in biofilms adopt a distinct phenotype that protects them from antimicrobial agents. This phenotype, however, is not due to starvation, but is programmed based on environmental factors (Costerton, 1999). This hypothesis is supported by studies that show resistance in new biofilms that are too thin to pose a physical barrier to antibiotic entrance. A major point is that all members of a biofilm must be removed or killed to prevent relapse, much in the same way that all cancer cells in a tumor must be removed or killed to prevent cancer reoccurrence. If as little as 1% of the original population persists after antimicrobial treatments, the infection can come back (Stewart, 2001). <br> | ||
<br> | <br> | ||
==Medical Treatments== | ==Medical Treatments== | ||
<br> | <br> | ||
Revision as of 23:22, 22 April 2015
Pseudomonas aeruginosa was first discovered and described as a distinct species by Pasture in the nineteenth century. It was studied as a pathogen, however, first by Carle Gessard, who noted that it turned bandages blue and green when exposed to ultra-violent radiation. Though outstanding advances in both microscopy and genetics, much more is currently known about this incredible species, which is extremely metabolically diverse, ubiquitous, and a devastating opportunistic pathogen.
To begin, P. aeruginosa is gram negative bacterium with one flagella (Lederber, 2000). The flagellum plays an extremely important role in the production and assembly of biofilm formation. It is also a facultative anaerobe, but its optimal metabolism is aerobic respiration. In the absence of oxygen, it can respire anaerobically using nitrate or an alternate electron acceptor(Costerton, 1999). However, it often cannot grow when it is simultaneously a pathogen and not exposed to oxygen, as is common in the depth of the biofilm. Many cells thus often remain in a un-dividing state when in a biofilm. This, however, works in P. aeruginosa’s favor, as many antibiotics cannot kill cells that are not actively growing or dividing (Stewart, 2001). Furthermore, it can catabolize a variety of organic molecules, even carcinogens such as benzoate. This incredible nutritional and metabolic versatility and diversity allows it to survive attack by both phage and human induced antimicrobial agents, as well as live in a diverse arrays of environments. It is found ubiquitously, in environments as diverse as hospitals, sewage, animals, plants, water, and the soil. It is also the most predominant organism in oligotrophic aquatic ecosystems. Furthermore, in the soil, P. aeruginosa is an important ecological bacteria, breaking down the polycyclic aromatic hydrocarbons, and catabolizing important molcuelcules such as rhamnolipids, quinolones, and hydrogen cyanide (Lederberg, 2000).This versatility allows it to be the most abundant organism on earth.
Despite this abundance, it is a pathogen. However, it only acts opportunistically, rarely infecting healthy individuals. It is common in immunocompromised patients, such as people suffering from AIDS or cancer (Lederberg, 2000). Cystic Fibrosis (CF) patients have an abundance of salt in their respiratory tract due to faulty CFTR chloride channels; this prevents the production and action of chemicals and proteins used by the immune system to prevent infection. As a result, many CF patients are chronically infected by P. aeruginosa, causing severe pulmonary inflammation and damage. As a result, the median life expectant of CF patients is 30 years old (Steward, 2001). Because of its prevalence in hospitals and selective pathogenesis, it infects two thirds of the critically ill hospitalized patients. Unfortunately, it is an extremely invasive pathogen, with a mortality rate of 40-60%. The mortality rate is extremely high because of the biofilm nature of P. aeruginosa. When a pathogen in the human lungs, it often forms biofilms, communities of bacterial cells attached to an inert or living surface and enclosed in a extracellular polysaccharide matrixes. These biofilms release planktonic bacteria, which are dispersed to spread the infection to other tissues or host organism. Many antibiotics kill the planktonic bacteria. However, antibiotics fail to kill the bacterium in the biofilm; for this reason, after antibiotic therapy is complete, the biofilm continues to produce planktonic bacteria and the infection reoccurs. Biofilms are inherently resistant to antibiotics, weather in the form of bacteriophages, amoebae, or chemical biocides (Lederberg, 2000).
Method of Pathology
P. aeruginosa is a ubiqoutous organism, found in many diverse envrinments. However, it is also an opportunistic pathogen. In hospitals, pathogenic biofilms have been found using electron microscopy on the surface of medical devices, such as central venous catheters, prosthetic heart valves, and orthopedic implants (Costerton, 1999; Stewart, 2001). P. Aeruginosa can be the only bacterial species in the biofilm, but it may comprise of a mixture of multiple bacterial and fungal infections (Costerton, 1999). Because of its prevalence in hospitals and selective pathogenesis, it infects two thirds of the critically ill hospitalized patients. Unfortunately, it is an extremely invasive pathogen, with a mortality rate of 40-60%. It is common in immunocompromised patients, such as people suffering from AIDS, cancer, or CF (Lederberg, 2000). The mortality rate is extremely high because of the biofilm nature of P. aeruginosa. When a pathogen in the human lungs, it often forms biofilms, communities of bacterial cells attached to an inert or living surface and enclosed in a extracellular polysaccharide matrixes. These biofilms release planktonic bacteria, which are dispersed to spread the infection to other tissues or host organism. Many antibiotics kill the planktonic bacteria. However, antibiotics fail to kill the bacterium in the biofilm; for this reason, after antibiotic therapy is complete, the biofilm continues to produce planktonic bacteria and the infection reoccurs. This biofilm grows slowly and disperses to multiple locations through planktonic bacteria. For this reason P. Aeruginosa infections often take a long time to produce overt symptoms. This type of infection often develops slowly. Only acute exacerbations, caused by Planktonic bacteria that have detached and dispersed from the original biofilm, can be cured by antibiotics. However, after antibiotic therapy is complete, the biofilm continues to produce planktonic bacteria and the infection reoccurs. For this reason, the only effective course of action is to surgically remove the biofilm if possible (Costerton, 1999).
When healthy individuals are infected by P. aeruginosa it is rarely resolved. Tissues near the biofilm may often become damaged by the excessive amount of neutrophils and immune complexes released in a healthy person’s immune response. This is the same reason the lungs of CF patients undergo much damage; the high number of antibodies produced often results in worsening health, as they cause inflammation and damage to pulmonary tissue (Stewart, 2001). For this reason, immunosuppression drugs are often given to CF patients— the innate immune system cannot fight off the biofilms and doing so just worsens health conditions. However, consistent treatment with antibiotics to prevent colonization in young patients is promising (Costerton, 1999).
Biofilms
Biofilms are organized communities of bacterial cells attached to an inert or living surface and enclosed in a extracellular polysaccharide matrixes. Biofilms were one of the first microbial communities to be studied in microbiology; Anton van Leeuwenhoek even scraped plaque biofilms from his teeth and observed the microbes with a rather primitive microscope (Costerton, 1999). The aforementioned matrix forms a slippery, solid coat around the bacterial community. These bacterial communities cause many persistent and chronic bacterial infections, such as P. Aeruginosa infection in immunocompromised patients. Studies of biofilms have shown that they often consist of differentiated cells that often work together. Biofilms also often are comprised of multiple species. An understanding of both biofilm production and interaction could provide new therapeutic targets (Costerton, 1999).
Development
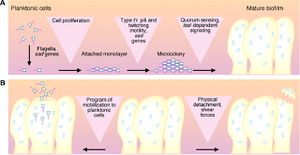
Biofilms work to protect the bacterial in a hostile environment, such as the human lungs. The cells differentiate, for example into structures that circulate nutrients. For this reason, cells in different regions of a biofilm exhibit distinctive and different patterns of gene expression (Costerton, 1999). The pattern of P. aeruginosa development is that preexisting P. aeruginosa release planktonic bacterial that disperse and multiply, spreading infection to other organisms or other tissues of the body. These planktonic bacterial than attach to a solid surface, form micro colonies on the surface, and differentiate into exoploysaccrahide biofilms. Planktonic cells then detach and disperse, restarting the cycle. Many antibiotics kill the planktonic bacteria released, but fail to kill the bacterium in the biofilm; for this reason, after antibiotic therapy is complete, the biofilm continues to produce planktonic bacteria and the infection reoccurs. One of the only ways to stop biofilm bacterial reproduction is to surgically remove the biofilm (Costerton, 1999).
The initial attachment requires both flagella, as show by an experiment done by O’Toole and Kolter, where knock out mutants without flagella could not adequately adhere to a surface. During this attachment phase, the transcription of particular genes is activated, such as algC, algD, and algU, which are required for the synthesis of the extracellular polysaccharide that encases the biofilm. This is spectacular because it implies that P. aeruginosa have a ‘sense of touch’ that enables the detection and reaction to a surface (Costerton, 1999). Next, Microcolonies are formed. Microcolonies are relatively small groups of bacteria. The formation of microcolonies requires Type IV pili, as shown by O’Toole and Kolter. Bacteria lacking pili were able to form a monolayer, but not microcolonies. These pili are involved in a ‘twitching,’ which is thought to be necessary to the aggregation of cells into microcolonies. These microcolonies then differentiate into true biofilms. Quorum sensing, the exchange of chemical signals between cells, may affect the timing of this differentiation as well as the coordinated building of the biofilm into complex structures. Finally, the planktonic cells detach and differentiate from the biofilms. One mechanism for this to occur is that the pieces of biofilms break off, but this break may be regulated based on molecular cues. For example, it is thought that the release of Planktonic cells requires an enzyme to digest alginate (Costerton, 1999).
Iron and Biofilm Production
Iron is a signal for biofilm production in P. aeruginosa. It is believed that either the level of internal iron of the active transport of chelated iron acts as a signal to produce biofilm formation. Iron starvation can often prevent the growth of bacteria, but sufficient levels may serve as a signal for biofilm development. This has been demonstrated by Lactoferrin, which sequesters iron and prevents P. aeruginosa from maturing from thin monolayers into multicellular biofilm structures. This could be die to the fact that low iron causes continuous twitching movements, the control of which is needed to form biofilm structures. Iron is often acquired by pyverdine, which is regulated by PvdS. When pyverdine binds to an outer-membrane receptor, it results in a signal the governs not only the activity of PvdS, but extracellular virulence factors. This is important for the opportunistic nature of P. aeruginosa infections; only when iron concentration is high and the cell is tethered to a surface, can pathogenic biofilm production begin. The Fur protein also regulates iron-responsive genes through a pair of small regulatory RNAs. The authors found that any pathway that allows iron into the cell –pryverdine or Fur, for example— can function as an iron-signaling pathway (and thus as a signal for virulence and biofilm development pathway). This indicates that a critical content of intracellular iron is a single for biofilm pathway initiation (Banin, 2005).
Gene Expression
P. aeruginosa planktonic cells and biofilm cells have extremely different lifestyles. Planktonic cells are not surrounded by an extracellular polysaccharide matrixes, like their biofilm enclosed counterparts, but instead dissociation and diffuse from the biofilm in order to spread the bacteria to new tissues and host organisms. Because of this, it is surprising that they two types of P. aeruginosa cells differ only by 1% in terms of differential gene expression. This refutes the argument that P. aeruginosa’s existence in a biofilm is dependent on dramatic difference in gene expression. However, the small amount of genes that are activated and repressed (34 and 39 respectively) may be crucial for the aggregation of the cells into a biofilm. Gene specifying pili and flagella are suppressed in biofilm bacteria, suggest that they are not needed for the incorporation and upkeep of mature biofilm. They are instead probably important only in the initial processed of biofilm development. Furthermore, the most activated gene in the biofilm P. aeruginosa were those from a bacteriophage related to Pf1. Pf1 genes were activated 100 to 1000 times more in biofilm than planktonic bacteria. This shows that phage induction is important for gene transfer within biofilms. It is suspected to function either to exclude certain strains of P. aeruginosa from biofilms induction or to contain a gene specifying an important toxin (Whitely, 2001).
Quorum Sensing
Quorum sensing is thought to determine when microcolonies differentiate into true biofilms in P. aeruginosa and other Gram Negative bacteria. Individual cells produce acylhomoserine lactone signals; when a sufficient number of acylhomoserine producing cells are present, the signals accumulate and trigger the transcription and translation of a specific set of genes (Costerton, 1999). Another P. aeruginosa quorum sensing trait related to differentiation has been characterized. The LasR-LasI controlling the expression of extracellular virulence factors, as well as the Rh1R and Rh1I, which control the expression of many secondary metabolites, such as butyrylhomoserine. Mutants of this process do not differentiate (Costerton, 1999). It is also believed that biofilm production could be halted if the cell-to-cell signaling molecules are blocked or destroyed. This destruction of the biofilm could allow the host immune system (or the host immune system supplemented with traditional antibiotics) to destroy the P. aeruginosa cells (Costerton, 1999; Stewart, 2001).
Antibiotic Resistance
Often, bacteria avoid bactericide by antibiotic using efflux pumps, modifying enzymes, or targeted mutations. However, these mutations are not what cause antimicrobial protection in biofilms. Studies on genes encoding efflux pumps, for example, revealed that P. aeruginosa with multidrug efflux pumps, such as Mar operon or the MexAB-OprM resistance pump, had the same resistance to antibiotics when grown in biofilms as mutant that did not produce Mar or MexAB-OprM (Stewart, 2001). Opposing research on gene expression in biofilms, however, dhows that over ten genomic islands encode efflux pumps and are activated in the presence of antibiotics. This is interesting because it shows that multiple antibiotics, as well as drugs targeting the production and upkeep of biofilms, may be necessary to cure not only P. aeruginosa infections, but all chronic infections caused by bacterial (or joint bacterial and fungal infections) (Whitely, 2001). Many antibiotics kill the planktonic bacteria released, but fail to kill the bacterium in the biofilm; for this reason, after antibiotic therapy is complete, the biofilm continues to produce planktonic bacteria and the infection reoccurs. There even appears to be a programmed detachment of planktonic bacteria. Biofilms are inherently resistant to antibiotics, weather in the form of bacteriophages, amoebae, or chemical biocides. They have this resistance through multiple mechanisms. The fact that the planktonic bacteria are killed by antibiotics suggests that resistance of biofilms is not due to mobile genetic elements mutations (Costerton, 1999).
One method of resistance is due to the failure of the antibiotic to fully diffuse through the biofilm. The slow the diffusion of antibiotics, allowing the antimicrobial agents to be deactivate in the outer layers of the biofilm. Even reactive oxidants, such as hydrogen peroxide, which are produced by phagocytic cells cannot diffuse through the entire biofilm, preventing biofilm death (Costerton, 1999). However, measurements of antibiotic diffusion in vitro have shown that a select few antibiotics readily fuse throughout the biofilm matrix, which is mostly water. If these antimicrobial agents are deactivator, however the diffusion can be slowed down dramatically (Stewart, 2001).
A second hypothesis centers around spatial heterogeneity. Many of the cells exist in different metabolic states, meaning that at least of the cells are guaranteed to survive a metabolically directed attack. For example, some of the cells in the biofilm lack access to sufficient nutrients and therefore are slow growing; these slow growing bacteria are not extremely susceptible to microbial agents (Costerton, 1999). Anther example is that Oxygen is often completely consumed by the surface layers of a biofilm, creating anaerobic niches in the less exposed regions of the biofilm. These anaerobic niches are often unaffected by Aminoglycoside antibiotics. pH differences can also occur due to accumulation of waste products. This could prevent the action of an antibiotic. This accumulation of acidic waste products can often cause bacterial cells to enter a non-growing state. Antibiotics, like Penicillin, which targets cell wall synthesis, only works on growing bacteria. So, the accumulation of waster effectively stops the antibiotic from killing all of the bacterial in the biofilm, a necessity for recovery from pathogens. This hypothesis is supported by visualization of metabolically inactive zones within biofilms. Finally, the osmotic environment could be changed, causing an osmotic stress response. This response could change the amount of porins and cell envelope permeability, effectively stopping high numbers of antibiotics from gaining entrance (Stewart, 2001).
Thirdly, it is believed that some of the cells in biofilms adopt a distinct phenotype that protects them from antimicrobial agents. This phenotype, however, is not due to starvation, but is programmed based on environmental factors (Costerton, 1999). This hypothesis is supported by studies that show resistance in new biofilms that are too thin to pose a physical barrier to antibiotic entrance. A major point is that all members of a biofilm must be removed or killed to prevent relapse, much in the same way that all cancer cells in a tumor must be removed or killed to prevent cancer reoccurrence. If as little as 1% of the original population persists after antimicrobial treatments, the infection can come back (Stewart, 2001).
Medical Treatments
Current therapies
Current therapies include strong courses of antibiotics. Additionally, consistent treatment with antibiotics to prevent colonization in young CF patients is promising (Costerton, 1999).
Potential Therapies
The knowledge of the steps of biofilm production provide new targets to cure P. aeruginosa. As there are multiple antibiotic resistance methods that might work together, antibiofilm therapies may have to prevent the successful mechanism to be successful clinically. Furthermore, biofilms are often heterogeneous, existing in a broad spectrum of metabolic states and growth rates; this could prevent not only the adequate spread of antibiotics, but its overall efficiency. Finally, a small portion of the cells might differentiate into a highly protected phenotype. These three facts make it unlikely that any one antimicrobial agent would be able to kill all of the cells in the biofilm (Stewart, 2001).
Most antibiotics used currently are for growing cells, but new screes of existing antibiotics against non-growing or biofilm cells may yield antibiotics effective on non-growing cells in P. aeruginosa infections (Stewart, 2001). Furthermore, genes that mediate biofilm resistance are currently being identified. When this process is complete there gene products can be characterized. This characterization of gene products could be used to make antimicrobial agents that would increase the efficiency of already existing antibiotics in biofilm production (Stewart, 2001). Another strategy could be to create therapies that disrupt the multicellular structure of biofilms. If the multicellular structure of the biofilm is broken down and disintegrated, the host immune system (or the host immune system supplemented with traditional antibiotics) might be able to destroy the cells in much the same way that Planktonic cells are often killed by the immune system or antibiotics, such as Penicillin. This could be done several ways. One, the infected cells could be exposed to high amounts of enzymes that dissolve the biofilm matrix polymers. Enzymes that block biofilm matrix synthesis or interfere with quorum sensing required for biofilm formation are also potential targets. These types of therapies target the formation of multicellular structures; this is opposed to current antibiotics, which disrupt essential functions of individual cells (Stewart, 2001).
References
[1] Banin, E. Proceedings of the National Academy of Sciences - PNAS: Iron and Pseudomonas Aeruginosa Biofilm Formation. 102 Vol. The Academy, 08/02/2005. Web. 20 Apr. 2015.
[2] Costerton, J. W. Science (New York, N.Y.): Bacterial Biofilms: A Common Cause of Persistent Infections. 284 Vol. American Association for the Advancement of Science, 05/21/1999. Web. 20 Apr. 2015.
[3] Lederberg, Joshua et al. Pseudomonas. Encyclopedia of Microbiology. Second Edition. Volume 3. San Diego, 2000. p. 876-891.
[4] Stewart, Philip S. The Lancet (North American Edition): Antibiotic Resistance of Bacteria in Biofilms. 358 Vol. Little, Brown and Co, 07/2001. Web. 20 Apr. 2015.
[5] Whiteley, Marvin. Nature (London): Gene Expression in Pseudomonas Aeruginosa Biofilms. 413 Vol. Macmillan Journals Ltd., etc, 10/2001. Web. 20 Apr. 2015.
Authored for BIOL 238 Microbiology, taught by Joan Slonczewski, 2015, Kenyon College.
