Pyrobaculum aerophilum: Difference between revisions
No edit summary |
|||
| (12 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
{{Curated}} | |||
{{Biorealm Genus}} | {{Biorealm Genus}} | ||
| Line 5: | Line 6: | ||
===Higher order taxa=== | ===Higher order taxa=== | ||
Archaea; Crenarchaeota; Thermoprotei; Thermoproteales; Thermoproteaceae; Pyrobaculum | Archaea; Crenarchaeota; Thermoprotei; Thermoproteales; Thermoproteaceae; [[Pyrobaculum]] | ||
===Species=== | ===Species=== | ||
| Line 19: | Line 20: | ||
==Description and significance== | ==Description and significance== | ||
[[Image:Aerophilum2.jpg|frame|thumb|200px|right|''Pyrobaculum aerophilum'' - [http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=182387 | [[Image:Aerophilum2.jpg|frame|thumb|200px|right|''Pyrobaculum aerophilum'' - [http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=182387 PubMed.]]] | ||
''Pyrobaculum aerophilum'' is a rod-shaped hyperthermophilic archaeum that was first isolated from boiling marine water in Maronti Beach, Ischia, Italy. ''Pyrobaculum aerophilum'' derives its name from the Greek noun "aer" (air) and the Greek adjective "philos" (loving). It was found that the archaeum grew optimally at 100°C and at pH 7.0. Both organic and inorganic compounds served as substrates during aerobic and anaerobic respiration. However, growth was inhibited by elemental sulfur. When discovered, ''Pyrobaculum aerophilum'' resembled members from the genera ''Thermoproteus'' and ''Pyrobaculum'' because of its ability to transform into spherical bodies, which resemble golf balls. After its 16S rRNA was sequenced, the new archaeum displayed traits more characteristic of the genus ''Pyrobaculum'' and was therefore classified as ''Pyrobaculum aerophilum''. Most species in the genus ''Pyrobaculum'' cannot live in the presence of oxygen; however | ''Pyrobaculum aerophilum'' is a rod-shaped hyperthermophilic archaeum that was first isolated from boiling marine water in Maronti Beach, Ischia, Italy. ''Pyrobaculum aerophilum'' derives its name from the Greek noun "aer" (air) and the Greek adjective "philos" (loving). It was found that the archaeum grew optimally at 100°C and at pH 7.0. Both organic and inorganic compounds served as substrates during aerobic and anaerobic respiration. However, growth was inhibited by elemental sulfur. When discovered, ''Pyrobaculum aerophilum'' resembled members from the genera ''Thermoproteus'' and ''Pyrobaculum'' because of its ability to transform into spherical bodies, which resemble golf balls. After its 16S rRNA was sequenced, the new archaeum displayed traits more characteristic of the genus ''Pyrobaculum'' and was therefore classified as ''Pyrobaculum aerophilum''. Most species in the genus ''Pyrobaculum'' cannot live in the presence of oxygen; however ''Pyrobaculum aerophilum'' can interestingly use oxygen for growth. [2] | ||
''Pyrobaculum aerophilum'' cells were usually found to be 3 to 8 μm long and 0.6 μm wide. Very rarely, however, cells with a length of 20 μm appeared. Motility is achieved using monopolar flagellation with bundles up to eight flagella. Each flagella averaged a diameter of 10 nm. Cells would transform into their terminal spherical phase mainly in the stationary-growth phase, at high nitrate concentrations, or pH values exceeding 8.0. Even in their terminal | ''Pyrobaculum aerophilum'' cells were usually found to be 3 to 8 μm long and 0.6 μm wide. Very rarely, however, cells with a length of 20 μm appeared. Motility is achieved using monopolar flagellation with bundles of up to eight flagella. Each flagella averaged a diameter of 10 nm. Cells would transform into their terminal spherical phase mainly in the stationary-growth phase, at high nitrate concentrations, or pH values exceeding 8.0. Even in their terminal spherical phase, the cells can still enlarge or shrink depending on conditions. Colonies of ''Pyrobaculum aerophilum'' in their spherical phase exhibited greyish-yellow colonies with a harsh surface. Anaerobically conditioned cells in their normal rod-shaped phase exhibited a deep green color while aerobically grown cells in their normal rod-shaped phase exhibited a brownish yellow color. [2] | ||
''Pyrobaculum aerophilum'' grew in | ''Pyrobaculum aerophilum'' grew in media ranging from temperatures of 75 and 104°C. At the optimum temperature of growth, 100°C, the optimal doubling rate of the cells was observed to be 180 minutes. At the extreme temperature range of 75°C, the doubling rate of the cells dropped to a rate of approximiately 5 days. There was no difference in growth rate when the archaeum was cultured either in aerobic and anaerobic conditions. When living conditions were observed in different pH conditions, life was observed from a range of pH 5.8 to 9.0. The optimum rate of growth was observed at pH 7.0. However, when ''Pyrobaculum aerophilum'' was introduced in an environment exceeding pH 8.0, no growth was observed. Also, when the pH was lowered to 5.5 or exceeded 9.0, rapid cell lysis occurred. Different salt conditions were also observed for growth and life. Optimal salt conditions were observed at 1.5% NaCl. Also, growth of cells was observed when there were no traces of NaCl in the solution; however, once NaCl concentrations exceeded 3.6%, no growth was observed. [2] | ||
==Genome structure== | ==Genome structure== | ||
| Line 54: | Line 55: | ||
==Cell structure and metabolism== | ==Cell structure and metabolism== | ||
[[Image:Aerophilum.jpg|frame|thumb|100px|right|''Pyrobaculum aerophilum'' S-layer, spacer, and cytoplasmic membrane. [http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=182387 | [[Image:Aerophilum.jpg|frame|thumb|100px|right|''Pyrobaculum aerophilum'' S-layer, spacer, and cytoplasmic membrane. [http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=182387 Pubmed]]] | ||
''Pyrobaculum aerophilum'' has characteristics of both ''Thermoproteus'' and ''Pyrobaculum''. The cylindrical rod-shape, the formation of terminal spheres, and the S-layer shows a close relationship to ''Thermoproteus'' and ''Pyrobaculum''. However, the archaeum was unable to grow by sulfur respiration and growth was suppressed by the presence of elemental sulfur. The | ''Pyrobaculum aerophilum'' has characteristics of both ''Thermoproteus'' and ''Pyrobaculum''. The cylindrical rod-shape, the formation of terminal spheres, and the S-layer shows a close relationship to ''Thermoproteus'' and ''Pyrobaculum''. However, the archaeum was unable to grow by sulfur respiration and growth was suppressed by the presence of elemental sulfur. The archaeum was also found to grow by dissimilatory nitrate reduction, organic compounds, and thiosulfate. Interestingly, nitrate reduction has only been observed within the heterotrophic extreme halophiles. [2] | ||
''Pyrobaculum aerophilum'' have a cell wall exhibiting a S-layer stationed on top of a cytoplasmic membrane. The S-layer was 5 nm in length on average and was anchored to the cytoplasmic membrane by a inner elongated domain (a spacer) that was 25 nm in length on average. The S-layer was consistent with other members of the genus ''Pyrobaculum'' including: ''P. islandicum'', ''P. organotrophum'', and ''T.tenax.'' [2] | ''Pyrobaculum aerophilum'' have a cell wall exhibiting a S-layer stationed on top of a cytoplasmic membrane. The S-layer was 5 nm in length on average and was anchored to the cytoplasmic membrane by a inner elongated domain (a spacer) that was 25 nm in length on average. The S-layer was consistent with other members of the genus ''Pyrobaculum'' including: ''P. islandicum'', ''P. organotrophum'', and ''T.tenax.'' [2] | ||
| Line 66: | Line 67: | ||
''Pyrobaculum aerophilum'' is a hyperthermophilic microogranism and therefore resides in hot, anaerobic environments. Because of this, it is difficult to technically study and characterize the functions of hot microbial ecosystems. However, it is known fermentative microorganisms oxidize amino acids and sugars to carbon dioxide and short-chain acids (of which acetate is by far the most plentiful). Consequently, the successful dispensation of organic matter in these anaerobic environments requires the activity of acetate-degrading anaerobic microorganisms (''Pyrobaculum aerophilum''). [7] | ''Pyrobaculum aerophilum'' is a hyperthermophilic microogranism and therefore resides in hot, anaerobic environments. Because of this, it is difficult to technically study and characterize the functions of hot microbial ecosystems. However, it is known fermentative microorganisms oxidize amino acids and sugars to carbon dioxide and short-chain acids (of which acetate is by far the most plentiful). Consequently, the successful dispensation of organic matter in these anaerobic environments requires the activity of acetate-degrading anaerobic microorganisms (''Pyrobaculum aerophilum''). [7] | ||
The study of acetate is important because it is the most prolific organic acid in hot microbial ecosystems. Acetate may enter hot microbial ecosystems from hotter, sterile environments in which organic matter is hydrolyzed to acetate. Acetate metabolism in ''Pyrobaculum aerophilum'' was observed in | The study of acetate is important because it is the most prolific organic acid in hot microbial ecosystems. Acetate may enter hot microbial ecosystems from hotter, sterile environments in which organic matter is hydrolyzed to acetate. Acetate metabolism in ''Pyrobaculum aerophilum'' was observed in media in which nitrate was provided as an electron acceptor [2]. In addition to nitrate, ''Pyrobaculum aerophilum'' is capable of utilizing iron(III) as an electron acceptor. However, ''Pyrobaculum aerophilum'' did not grow with acetate as the sole electron donor and iron(III) as the electron acceptor. Fe(III) is believed to be accessible as an electron acceptor in hot, anaerobic ecosystems such as geysers and environments around hydrothermal vents. As a result, hyperthermophiles (''Pyrobaculum aerophilum'') and other acetate-oxidizing iron(III) reducers may function in the oxidation of compounds in hyperthermophilic environments in a way similar to that of the mesophilic food chains involved in handling organic matter. [7] | ||
==Pathology== | ==Pathology== | ||
''Pyrobaculum aerophilum'' has not been associated with the cause of any disease or impairment of health in humans | ''Pyrobaculum aerophilum'' has not been associated with the cause of any disease or impairment of health in humans or other animals. This is because ''Pyrobaculum aerophilum'' survives in conditions ranging from 74 to 104°C. In humans, average body temperature is 37°C. This is well outside the required living conditions of ''Pyrobaculum aerophilum.'' [6] | ||
==Application to Biotechnology== | ==Application to Biotechnology== | ||
''Pyrobaculum aerophilum'' utilize amylomaltases, which are 4-alpha-glucanotransferases of glycoside hydrolase family 77 that transfer alpha-1,4-linked glucans to an alternative acceptor. The amylomaltase-encoding gene of ''Pyrobaculum aerophilum'' has been cloned and expressed in Escherichia coli. The gene product (PyAMase) was characterized and was found to display peak activity at 95°C and pH 6.5. Amylomaltase was discovered to act on gelatinized food-grade potato starch, producing a thermoreversible starch product with gelatin-like properties. This thermoreversible gel has potential applications in the food industry. [8] | ''Pyrobaculum aerophilum'' utilize amylomaltases, which are 4-alpha-glucanotransferases of glycoside hydrolase family 77 that transfer alpha-1,4-linked glucans to an alternative acceptor. The amylomaltase-encoding gene of ''Pyrobaculum aerophilum'' has been cloned and expressed in ''Escherichia coli''. The gene product (PyAMase) was characterized and was found to display peak activity at 95°C and pH 6.5. Amylomaltase was discovered to act on gelatinized food-grade potato starch, producing a thermoreversible starch product with gelatin-like properties. This thermoreversible gel has potential applications in the food industry. [8] | ||
''Pyrobaculum aerophilum'' is being examined because of its ability to survive in extreme environments. Interestingly, ''Pyrobaculum aerophilum'' can grow aerobically, unlike most of its thermophilic relatives, making it acquiescent to a variety of experimental exploitations and a candidate as a model organism for studying archaeal and thermophilic microbiology. Projects have been initiated to extract and purify proteins for structure determination by NMR and x-ray diffraction. The resulting three-dimensional structure of ''Pyrobaculum aerophilum'' will provide the knowledge to understand and manipulate protein function. Additionally to being an ideal exemplary candidate, ''Pyrobaculuum aerophilum'' is heavily being analyzed because of its relationship to Crenarchaeaeocytes, which may be the eukaryote's closest prokaryotic kin. [9] | ''Pyrobaculum aerophilum'' is being examined because of its ability to survive in extreme environments. Interestingly, ''Pyrobaculum aerophilum'' can grow aerobically, unlike most of its thermophilic relatives, making it acquiescent to a variety of experimental exploitations and a candidate as a model organism for studying archaeal and thermophilic microbiology. Projects have been initiated to extract and purify proteins for structure determination by NMR and x-ray diffraction. The resulting three-dimensional structure of ''Pyrobaculum aerophilum'' will provide the knowledge to understand and manipulate protein function. Additionally to being an ideal exemplary candidate, ''Pyrobaculuum aerophilum'' is heavily being analyzed because of its relationship to Crenarchaeaeocytes, which may be the eukaryote's closest prokaryotic kin. [9] | ||
| Line 79: | Line 80: | ||
==Current Research== | ==Current Research== | ||
1. [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=17395198&query_hl=14&itool=pubmed_DocSum | 1. [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=17395198&query_hl=14&itool=pubmed_DocSum Discovery of a thermophilic protein complex stabilized by topologically interlinked chains.] | ||
In one study, ''Pyrobaculum aerophilum'' was isolated to study how cellular proteins retain their natural folds under extreme living conditions. Recent | In one study, ''Pyrobaculum aerophilum'' was isolated to study how cellular proteins retain their natural folds under extreme living conditions. Recent studies using structural and computational tools have discovered that disulfide bonding is a crucial instrument for stabilizing proteins in many thermophilic organisms. Isolates of ''Pyrobaculum aerophilum'' reveal that the use of disulfide bonds include multiple protein-protein complexes, contrary to beliefs that individual protein complexes might be responsible for the composition of the native folds. In one complex, a citrate synthase homodimer contained two intramolecular disulfide bonds, which consequently led to a cyclization of each protein. This cyclization interlinked the two proteins and caused them to be inseparable. These protein interactions highlight the astonishing types of mechanisms created by evolution. [3] | ||
2. [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=17394648&query_hl=14&itool=pubmed_DocSum | 2. [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=17394648&query_hl=14&itool=pubmed_DocSum Phylogenomic analysis of proteins that are distinctive of Archaea and its main subgroups and the origin of methanogenesis.] | ||
Even today, a minute amount of molecular characteristics that are communal within all Archaea are recognized. In addition, many of the relationships between diverse groups within the Euryarchaeota are not openly understood. Thorough examinations on open reading frames in the genomes of 11 Archaea have been performed to explore proteins that are exclusive to all Archaea. The 11 archaea studied include: 3 Crenarchaeota - ''Aeropyrum pernix'', ''Pyrobaculum aerophilum,'' and ''Sulfolobus acidocaldarius''; 8 Euryarchaeota - ''Pyrococcus abyssi'', ''Methanococcus maripaludis'', ''Methanopyrus kandleri'', ''Methanococcoides burtonii'', ''Halobacterium sp. NCR-1'', ''Haloquadratum walsbyi'', ''Thermoplasma acidophilum'', and ''Picrophilus torridus''. The analyses of the open reading frames detected 1448 proteins that are unique to Archaea. Of the 1448 proteins, 6 of the proteins were unique to all Archaea, 10 others were missing only in Nanoarchaeum equitans, and a large number of other proteins were specific for various main groups within the Archaea. On the same note, 31 proteins were nearly present in all methanogens and 10 supplementary proteins were found only in select methanogens. The identified Archaea-specific proteins offer new molecular markers that are distinctive characteristics of Archaea and all of its main subgroups. The species distributions of these proteins provide new insights into the evolutionary relationships among different groups within Archaea, particularly regarding the origin of methanogenesis. [4] | Even today, a minute amount of molecular characteristics that are communal within all Archaea are recognized. In addition, many of the relationships between diverse groups within the Euryarchaeota are not openly understood. Thorough examinations on open reading frames in the genomes of 11 Archaea have been performed to explore proteins that are exclusive to all Archaea. The 11 archaea studied include: 3 Crenarchaeota - ''Aeropyrum pernix'', ''Pyrobaculum aerophilum,'' and ''Sulfolobus acidocaldarius''; 8 Euryarchaeota - ''Pyrococcus abyssi'', ''Methanococcus maripaludis'', ''Methanopyrus kandleri'', ''Methanococcoides burtonii'', ''Halobacterium sp. NCR-1'', ''Haloquadratum walsbyi'', ''Thermoplasma acidophilum'', and ''Picrophilus torridus''. The analyses of the open reading frames detected 1448 proteins that are unique to Archaea. Of the 1448 proteins, 6 of the proteins were unique to all Archaea, 10 others were missing only in ''Nanoarchaeum equitans'', and a large number of other proteins were specific for various main groups within the Archaea. On the same note, 31 proteins were nearly present in all methanogens and 10 supplementary proteins were found only in select methanogens. The identified Archaea-specific proteins offer new molecular markers that are distinctive characteristics of Archaea and all of its main subgroups. The species distributions of these proteins provide new insights into the evolutionary relationships among different groups within Archaea, particularly regarding the origin of methanogenesis. [4] | ||
3. [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=17085561&query_hl=14&itool=pubmed_DocSum | 3. [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=17085561&query_hl=14&itool=pubmed_DocSum CC1, a novel crenarchaeal DNA binding protein.] | ||
Single-stranded DNA binding proteins are crucial for DNA replication, recombination, and repair. Therefore, single-stranded DNA binding proteins are found virtually across the three domains of life. However, the genomes of the Crenarchaea ''Pyrobaculum aerophilum'' and ''Thermoproteus tenax'' are deficient in any noticeable single-stranded DNA binding proteins. In addition, these two genomes possess only one identifiable gene encoding a chromatin protein. The majority of Archaea have at least two diverse chromatin proteins. Biochemical tests for new nucleic binding proteins were carried out in cell extracts of ''Thermoproteus tenax''. Results identified three proteins capable of binding to single-stranded DNA. One protein, Alba, has been previously identified and is known to attach to single-stranded and duplex DNA. The other two proteins, delegated CC1, were found to be related to each other. These proteins were found to bind single and duplex DNA with similar | Single-stranded DNA binding proteins are crucial for DNA replication, recombination, and repair. Therefore, single-stranded DNA binding proteins are found virtually across the three domains of life. However, the genomes of the Crenarchaea ''Pyrobaculum aerophilum'' and ''Thermoproteus tenax'' are deficient in any noticeable single-stranded DNA binding proteins. In addition, these two genomes possess only one identifiable gene encoding a chromatin protein. The majority of Archaea have at least two diverse chromatin proteins. Biochemical tests for new nucleic binding proteins were carried out in cell extracts of ''Thermoproteus tenax''. Results identified three proteins capable of binding to single-stranded DNA. One protein, Alba, has been previously identified and is known to attach to single-stranded and duplex DNA. The other two proteins, delegated CC1, were found to be related to each other. These proteins were found to bind single and duplex DNA with similar affinity. Even in the absence of single-stranded DNA binding proteins, evolution has bestowed these organisms with other machinery to carry out the life-sustaining processes of replication, recombination, and repair. [5] | ||
==References== | ==References== | ||
[http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=11792869&dopt=Abstract | 1. [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=11792869&dopt=Abstract Fitz-Gibbon ST, Ladner H, Kimm UJ, Stetter KO, Simon MI, and Miller JH. "Genome Sequence of the hyperthermophilic crenarchaeon Pyrobaculum aerophilum." ''Proceedings of the National Academy of Sciences'', 2002 Jan. 22; 99(2) 984-9.] | ||
[http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=182387 | 2. [http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=182387 P Völkl, R Huber, E Drobner, R Rachel, S Burggraf, A Trincone, and K O Stetter. "Pyrobaculum aerophilum sp. nov., a Novel Nitrate-Reducing Hyperthermophilic Archaeum." ''Applied and Environmental Microbiology, Sept. 1993, p. 2918-2926; 0099-2240/93/092918.] | ||
[http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=17395198&query_hl=14&itool=pubmed_DocSum | 3. [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=17395198&query_hl=14&itool=pubmed_DocSum Boutz DR, Cascio D, Whitelegge J, Perry LJ, Yeates TO. "Discovery of a thermophilic protein complex stabilized by topologically interlinked chains." ''Journal of Molecular Biology.'' 2007 May 18;368(5):1332-44. Epub 2007 Mar 6.] | ||
[http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=17394648&query_hl=14&itool=pubmed_DocSum | 4. [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=17394648&query_hl=14&itool=pubmed_DocSum Gao B, Gupta RS. "Phylogenomic analysis of proteins that are distinctive of Archaea and its main subgroups and the origin of methanogenesis." ''BMC Genomics.'' 2007 Mar 29;8:86.] | ||
[http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=17085561&query_hl=14&itool=pubmed_DocSum | 5. [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=17085561&query_hl=14&itool=pubmed_DocSum Luo X, Schwarz-Linek U, Botting CH, Hensel R, Siebers B, White MF. "CC1, a novel crenarchaeal DNA binding protein." ''Journal of Bacteriology'', 2007 Jan;189(2):403. Epub 2006 Nov 3.] | ||
[http://genamics.com/cgi-bin/genamics/genomes/genomesearch.cgi?field=ID&query=1430 | 6. [http://genamics.com/cgi-bin/genamics/genomes/genomesearch.cgi?field=ID&query=1430 "Pyrobaculum Aerophilum." Genamics. 2006. GOLD: Genomes Online Database. 4 June 2007 <http://genamics.com/cgi-bin/genamics/genomes/genomesearch.cgi?field=ID&query=1430>.] | ||
[http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=92735 | 7. [http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=92735 Jason M. Tor, Kazem Kashefi, and Derek R. Lovley. "Acetate Oxidation Coupled to Fe(III) Reduction in Hyperthermophilic Microorganisms." Applied and Environmental Microbiology. 2001 March; 67(3): 1363–1365.] | ||
[http://aem.asm.org/cgi/content/abstract/71/9/5098 | 8. [http://aem.asm.org/cgi/content/abstract/71/9/5098 Thijs Kaper, Boguslawa Talik, Thijs J. Ettema, Herman Bos, Marc J. E. C. van der Maarel, and Lubbert Dijkhuizen. "Amylomaltase of Pyrobaculum aerophilum IM2 Produces Thermoreversible Starch Gels." Applied and Environmental Microbiology. September 2005, p. 5098-5106, Vol. 71, No. 9.] | ||
9. [http://www.ornl.gov/sci/techresources/Human_Genome/publicat/microbial/mgp.pdf Melvin I. Simon, Sorel Fitz-Gibbon. "Whole-Genome Sequence of ''Pyrobaculum aerophilum.'' Microbial Genome Program. 2000 February; p. 41-42.] | |||
Edited by a student of [mailto:ralarsen@ucsd.edu Rachel Larsen] and Kit Pogliano. | Edited by a student of [mailto:ralarsen@ucsd.edu Rachel Larsen] and Kit Pogliano. | ||
KMG | |||
Latest revision as of 15:06, 7 July 2011
A Microbial Biorealm page on the genus Pyrobaculum aerophilum
Classification
Higher order taxa
Archaea; Crenarchaeota; Thermoprotei; Thermoproteales; Thermoproteaceae; Pyrobaculum
Species
Pyrobaculum aerophilum
|
NCBI: Taxonomy |
Description and significance
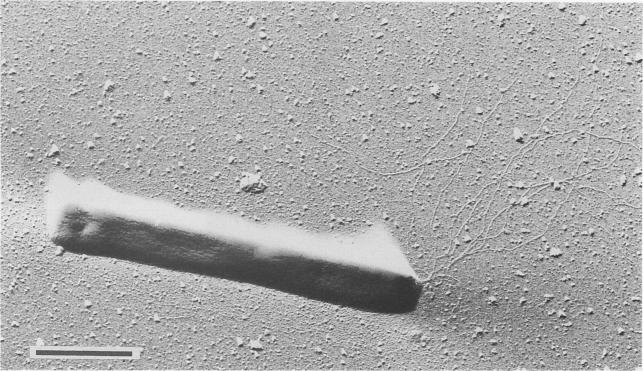
Pyrobaculum aerophilum is a rod-shaped hyperthermophilic archaeum that was first isolated from boiling marine water in Maronti Beach, Ischia, Italy. Pyrobaculum aerophilum derives its name from the Greek noun "aer" (air) and the Greek adjective "philos" (loving). It was found that the archaeum grew optimally at 100°C and at pH 7.0. Both organic and inorganic compounds served as substrates during aerobic and anaerobic respiration. However, growth was inhibited by elemental sulfur. When discovered, Pyrobaculum aerophilum resembled members from the genera Thermoproteus and Pyrobaculum because of its ability to transform into spherical bodies, which resemble golf balls. After its 16S rRNA was sequenced, the new archaeum displayed traits more characteristic of the genus Pyrobaculum and was therefore classified as Pyrobaculum aerophilum. Most species in the genus Pyrobaculum cannot live in the presence of oxygen; however Pyrobaculum aerophilum can interestingly use oxygen for growth. [2]
Pyrobaculum aerophilum cells were usually found to be 3 to 8 μm long and 0.6 μm wide. Very rarely, however, cells with a length of 20 μm appeared. Motility is achieved using monopolar flagellation with bundles of up to eight flagella. Each flagella averaged a diameter of 10 nm. Cells would transform into their terminal spherical phase mainly in the stationary-growth phase, at high nitrate concentrations, or pH values exceeding 8.0. Even in their terminal spherical phase, the cells can still enlarge or shrink depending on conditions. Colonies of Pyrobaculum aerophilum in their spherical phase exhibited greyish-yellow colonies with a harsh surface. Anaerobically conditioned cells in their normal rod-shaped phase exhibited a deep green color while aerobically grown cells in their normal rod-shaped phase exhibited a brownish yellow color. [2]
Pyrobaculum aerophilum grew in media ranging from temperatures of 75 and 104°C. At the optimum temperature of growth, 100°C, the optimal doubling rate of the cells was observed to be 180 minutes. At the extreme temperature range of 75°C, the doubling rate of the cells dropped to a rate of approximiately 5 days. There was no difference in growth rate when the archaeum was cultured either in aerobic and anaerobic conditions. When living conditions were observed in different pH conditions, life was observed from a range of pH 5.8 to 9.0. The optimum rate of growth was observed at pH 7.0. However, when Pyrobaculum aerophilum was introduced in an environment exceeding pH 8.0, no growth was observed. Also, when the pH was lowered to 5.5 or exceeded 9.0, rapid cell lysis occurred. Different salt conditions were also observed for growth and life. Optimal salt conditions were observed at 1.5% NaCl. Also, growth of cells was observed when there were no traces of NaCl in the solution; however, once NaCl concentrations exceeded 3.6%, no growth was observed. [2]
Genome structure
Pyrobaculum aerophilum is composed of a 2.2-megabase genome sequence. A whole genome analysis was computerized and experiments have confirmed that Pyrobaculum aerophilum and possibly all Crenarchaea lack 5’ untranslated regions in their mRNAs, and therefore do not appear to use a ribosome binding site. The lengths and dispersion of mononucleotide repeat-tracts uncovered that the Gs and Cs are highly precarious. This suggests that the archaeum could be lacking in a mismatch repair system. [1]
Complete Genome Sequence, completed 12/12/2001 @ UCLA/CalTech
Summary: Length: 2,222,430 nt
GC Content: 51%
% Coding: 88%
Topology: circular
Molecule: dsDNA
Genes: 2706
Protein coding: 2605
Structural RNAs: 96
Pseudo genes: none
Cell structure and metabolism
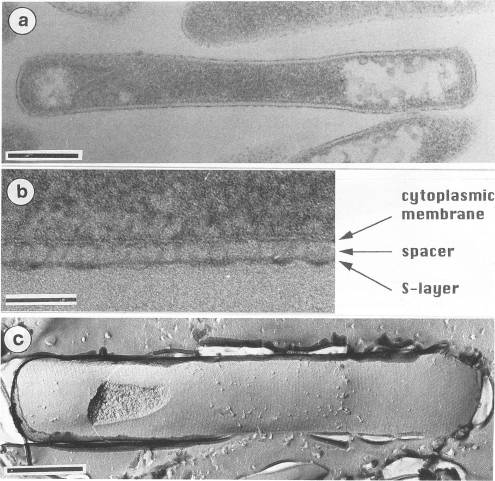
Pyrobaculum aerophilum has characteristics of both Thermoproteus and Pyrobaculum. The cylindrical rod-shape, the formation of terminal spheres, and the S-layer shows a close relationship to Thermoproteus and Pyrobaculum. However, the archaeum was unable to grow by sulfur respiration and growth was suppressed by the presence of elemental sulfur. The archaeum was also found to grow by dissimilatory nitrate reduction, organic compounds, and thiosulfate. Interestingly, nitrate reduction has only been observed within the heterotrophic extreme halophiles. [2]
Pyrobaculum aerophilum have a cell wall exhibiting a S-layer stationed on top of a cytoplasmic membrane. The S-layer was 5 nm in length on average and was anchored to the cytoplasmic membrane by a inner elongated domain (a spacer) that was 25 nm in length on average. The S-layer was consistent with other members of the genus Pyrobaculum including: P. islandicum, P. organotrophum, and T.tenax. [2]
Pyrobaculum aerophiluum is amazing in that it can utilize five different oxidants in respiration: nitrate, oxygen, arsenate, selenate, iron (III), and thiosulfate. During growth by nitrate respiration, nitrite and nitric oxide buildup was observed. However, no traces of ammonia was observed. When Pyrobaculum aerophilum was isolated with nitrite as an electron acceptor, no growth was obtained unless the nitrite was mixed with the presence of organic substrates. The products of nitrite reduction with the presence of organic substrates were nitrous oxide and nitric oxide. In conditions where both nitrate and nitrite were present, nitrate was completely consumed before nitrite was utilized. When growth with sulfur was diagnosed, an immediate inhibition of growth was observed and led to the eventual lysis of the cells. [2]
Ecology
Pyrobaculum aerophilum is a hyperthermophilic microogranism and therefore resides in hot, anaerobic environments. Because of this, it is difficult to technically study and characterize the functions of hot microbial ecosystems. However, it is known fermentative microorganisms oxidize amino acids and sugars to carbon dioxide and short-chain acids (of which acetate is by far the most plentiful). Consequently, the successful dispensation of organic matter in these anaerobic environments requires the activity of acetate-degrading anaerobic microorganisms (Pyrobaculum aerophilum). [7]
The study of acetate is important because it is the most prolific organic acid in hot microbial ecosystems. Acetate may enter hot microbial ecosystems from hotter, sterile environments in which organic matter is hydrolyzed to acetate. Acetate metabolism in Pyrobaculum aerophilum was observed in media in which nitrate was provided as an electron acceptor [2]. In addition to nitrate, Pyrobaculum aerophilum is capable of utilizing iron(III) as an electron acceptor. However, Pyrobaculum aerophilum did not grow with acetate as the sole electron donor and iron(III) as the electron acceptor. Fe(III) is believed to be accessible as an electron acceptor in hot, anaerobic ecosystems such as geysers and environments around hydrothermal vents. As a result, hyperthermophiles (Pyrobaculum aerophilum) and other acetate-oxidizing iron(III) reducers may function in the oxidation of compounds in hyperthermophilic environments in a way similar to that of the mesophilic food chains involved in handling organic matter. [7]
Pathology
Pyrobaculum aerophilum has not been associated with the cause of any disease or impairment of health in humans or other animals. This is because Pyrobaculum aerophilum survives in conditions ranging from 74 to 104°C. In humans, average body temperature is 37°C. This is well outside the required living conditions of Pyrobaculum aerophilum. [6]
Application to Biotechnology
Pyrobaculum aerophilum utilize amylomaltases, which are 4-alpha-glucanotransferases of glycoside hydrolase family 77 that transfer alpha-1,4-linked glucans to an alternative acceptor. The amylomaltase-encoding gene of Pyrobaculum aerophilum has been cloned and expressed in Escherichia coli. The gene product (PyAMase) was characterized and was found to display peak activity at 95°C and pH 6.5. Amylomaltase was discovered to act on gelatinized food-grade potato starch, producing a thermoreversible starch product with gelatin-like properties. This thermoreversible gel has potential applications in the food industry. [8]
Pyrobaculum aerophilum is being examined because of its ability to survive in extreme environments. Interestingly, Pyrobaculum aerophilum can grow aerobically, unlike most of its thermophilic relatives, making it acquiescent to a variety of experimental exploitations and a candidate as a model organism for studying archaeal and thermophilic microbiology. Projects have been initiated to extract and purify proteins for structure determination by NMR and x-ray diffraction. The resulting three-dimensional structure of Pyrobaculum aerophilum will provide the knowledge to understand and manipulate protein function. Additionally to being an ideal exemplary candidate, Pyrobaculuum aerophilum is heavily being analyzed because of its relationship to Crenarchaeaeocytes, which may be the eukaryote's closest prokaryotic kin. [9]
Current Research
1. Discovery of a thermophilic protein complex stabilized by topologically interlinked chains.
In one study, Pyrobaculum aerophilum was isolated to study how cellular proteins retain their natural folds under extreme living conditions. Recent studies using structural and computational tools have discovered that disulfide bonding is a crucial instrument for stabilizing proteins in many thermophilic organisms. Isolates of Pyrobaculum aerophilum reveal that the use of disulfide bonds include multiple protein-protein complexes, contrary to beliefs that individual protein complexes might be responsible for the composition of the native folds. In one complex, a citrate synthase homodimer contained two intramolecular disulfide bonds, which consequently led to a cyclization of each protein. This cyclization interlinked the two proteins and caused them to be inseparable. These protein interactions highlight the astonishing types of mechanisms created by evolution. [3]
Even today, a minute amount of molecular characteristics that are communal within all Archaea are recognized. In addition, many of the relationships between diverse groups within the Euryarchaeota are not openly understood. Thorough examinations on open reading frames in the genomes of 11 Archaea have been performed to explore proteins that are exclusive to all Archaea. The 11 archaea studied include: 3 Crenarchaeota - Aeropyrum pernix, Pyrobaculum aerophilum, and Sulfolobus acidocaldarius; 8 Euryarchaeota - Pyrococcus abyssi, Methanococcus maripaludis, Methanopyrus kandleri, Methanococcoides burtonii, Halobacterium sp. NCR-1, Haloquadratum walsbyi, Thermoplasma acidophilum, and Picrophilus torridus. The analyses of the open reading frames detected 1448 proteins that are unique to Archaea. Of the 1448 proteins, 6 of the proteins were unique to all Archaea, 10 others were missing only in Nanoarchaeum equitans, and a large number of other proteins were specific for various main groups within the Archaea. On the same note, 31 proteins were nearly present in all methanogens and 10 supplementary proteins were found only in select methanogens. The identified Archaea-specific proteins offer new molecular markers that are distinctive characteristics of Archaea and all of its main subgroups. The species distributions of these proteins provide new insights into the evolutionary relationships among different groups within Archaea, particularly regarding the origin of methanogenesis. [4]
3. CC1, a novel crenarchaeal DNA binding protein.
Single-stranded DNA binding proteins are crucial for DNA replication, recombination, and repair. Therefore, single-stranded DNA binding proteins are found virtually across the three domains of life. However, the genomes of the Crenarchaea Pyrobaculum aerophilum and Thermoproteus tenax are deficient in any noticeable single-stranded DNA binding proteins. In addition, these two genomes possess only one identifiable gene encoding a chromatin protein. The majority of Archaea have at least two diverse chromatin proteins. Biochemical tests for new nucleic binding proteins were carried out in cell extracts of Thermoproteus tenax. Results identified three proteins capable of binding to single-stranded DNA. One protein, Alba, has been previously identified and is known to attach to single-stranded and duplex DNA. The other two proteins, delegated CC1, were found to be related to each other. These proteins were found to bind single and duplex DNA with similar affinity. Even in the absence of single-stranded DNA binding proteins, evolution has bestowed these organisms with other machinery to carry out the life-sustaining processes of replication, recombination, and repair. [5]
References
Edited by a student of Rachel Larsen and Kit Pogliano.
KMG
