Pyrolobus fumarii: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
{{Uncurated}} | |||
{{Biorealm Genus}} | {{Biorealm Genus}} | ||
Latest revision as of 03:32, 20 August 2010
A Microbial Biorealm page on the genus Pyrolobus fumarii
Classification
Higher order taxa:
Archaea; Crenarchaeota; Thermoprotei; Desulfurococcales; Pyrodictiaceae
Species:
Pyrolobus fumarii (Synonyms: Pyrolobus Blochl et al. 1999)
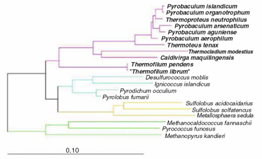
Phylogenetic Tree
Description and Significance
Pyrolobus fumarii are made up of cells that are regularly to irregularly lobed cocci. Chemolithoautotrophic growth by anaerobic and microaerophilic H2 oxidation with NO3-, S2O32-, and O2 as electron accepters. By doing so well at temperatures of 95ºC to 113ºC, our knowledge about the upper limits of life is expanded. (Huber et al. 2006)
Genome Structure
The genome of P. fumarii is expected to contain many novel metabolic enzymes of commercial interest because it is able to survive on inorganic chemicals, carbon dioxide, and hydrogen. The genome is 1.85 million base pairs in length and contains approximately 2,000 genes. This organism, at first glance, seems to have an unusually high number of genes that have no obvious similarity to previously described genes from archaea and eubacteria (two major classifications of life). (Diversa 2001)
DNA Base composition Determined by a direct analysis of its mononucleosides and melting point analysis,Pyrolobus fumarii has a cytosine-guanine composition of 52.9mol% and 53.4mol% respectively. (Blöchl et al. 1997)
DNA-DNA homology The closest relatives of Pyrolobus fumarii are members of Pyrodictium based off 16S rRNA partial sequencing. There is insignificant DNA homology between Pyrolobus fumarii, Pyrodictium occultum, and Pyrodictium abyssi as shown in a DNA/DNA hybridization. (Blöchl et al. 1997)
Cell Structure
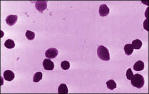
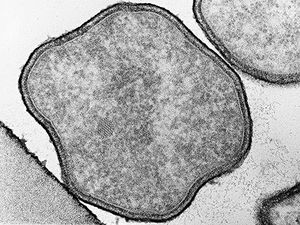
Pyrolobus fumarii are cells are regularly to irregularly lobe-shaped cocci (0.7-2.5 microm in diameter) arranged singly. A protein surface layer composes the cell envelope, which exhibits p4 symmetry. (Goncalves et al. 2008)
Metabolism
Pyrolobus fumarii is a facultatively aerobic obligate chemolithoautotroph. Moreover, P. fumarii is hydrogen – dependent gaining energy by H2 oxidation at a very low oxygen concentration and uses CO2¬ as the single carbon source. (Diversa 2001)
There are three different metabolic types observed on electron acceptor dependence:
- Nitrate Ammonification
- Nitrate is used as a terminal acceptor during strict anaerobic conditions wherein NO3- and H2 are present. In the process, ammonia is formed during NO3- reduction.
- Thiosulfate reduction
- Formation of H2S in the presence of H2 and uses S2O32- as electron acceptor also during anaerobic conditions.
- Microaerophilic hydrogen oxidation
- Under increased O2 condition, H2 and O2 were the only contributing elements for growth. In this reaction, O2 serves as the electron acceptor.
In addition, acetate, pyruvate, glucose, starch, or elemental sulfur are capable of inhibiting the growth of P. fumarii and these organic compounds do not contribute in growth stimulation. P. fumarii isolates elicited hydrogenase activity. It used methylene blue as the artificial electron acceptor and the process was membrane –associated. To allow existence in aggressive conditions, heat shock proteins were also detected in P. fumarii. Isolates were observed to have a chaperone-like heat shock protein. Recent study (Gonc¸alves et. al.) shows that the major organic solute composition of P. fumarii is DIP or Di-myo-inositol phosphate. The accumulation of DIP is considered to be one of the metabolic features of P. fumarii enabling it to survive as hyperthermophile living at the upper temperature and pressure limits for life. Metabolic features of Pyrolobus suggest its capability to exist in a pre-Earth condition with solely volcanic activity and liquid water conditions. (Diversa 2001)
Ecology
Pyrolobus fumarii is an extreme hyperthermophile isolated in the wall of a black smoker, or a deep-sea hydrothermal vent formed by sulfur-bearing minerals from beneath the Earth's crust, located in the Mid-Atlantic Ridge. It thrives at temperature of superheated water with optimal temperature at 106ºC and will not grow below 90ºC. Its optimum pH is 5.5 and grew between 4.0 – 6.5 while salt concentration is between 1%-4% with 1.7% as its optimum. (Diversa 2001)
Other Importance
Pyrolobus fumarii has other applications such as agricultural product processing enzymes, animal-feed additives, and industrial and consumer product enzymes. Applications for such highly thermostable products include many animal-feed additives, agricultural product processing enzymes, and industrial and consumer product enzymes. (Diversa 2001)
References
Blöchl, E., R. Rachel, S. Burggraf, D. Hafenbradl, H.W. Jannasch, and K.O. Stetter. 1997. Pyrolobus fumarii, gen. and sp. nov., represents a novel group of archaea, extending the upper temperature limit for life to 113ºC. Exptremeophiles 1:14-27.
Diversa Corporation. 25 September 2001. Diversa Completes of Sequencing of Pyrolobus fumarii Genome - the World's Highest Temperature Organism. 10 April 2008. <http://www.spaceref.com/news/viewpr.html?pid=6101>.
Goncalves, L.G., P. Lamosa, R. Huber, H. Santos. 2008. Di-myo-inositol phosphate and novel UDP-sugars accumulate in the extreme hyperthermophile Pyrolobus fumarii. Extremophiles.
Huber, H., R. Huber, and K.O. Stetter. 2006. Thermoproteales. Prokaryotes 3:10-22.