Rhodopseudomonas palustris and its role in bioremediation of soil pollutants
Introduction
Rhodopseudomonas palustris belongs to the group of phototrophic non-sulfur purple bacteria. It is a gram negative bacterium commonly found in soil and water. It can survive in both aerobic and anaerobic conditions. It is considered to be a “versatile bacteria” because it is able to fix carbon dioxide to form biomass, convert nitrogen to ammonia and produce hydrogen(hydrogen gas being the byproduct of nitrogen fixation)[1]. In the presence of oxygen it catabolizes a range of hydrocarbons from sugars to lignin; however it can switch to photosynthesis in the absence of oxygen. It is increasingly gaining research interest due to its abilities to produce hydrogen and degrade toxic waste. These two have very important applications in bioremediation and alternative energy.
It is in the genes
Caroline Harwood, who serves as the editor of the journal
Applied and Environmetnal Microbiolgy, lead the project that sequenced the genome of
R. palustris. Harwood started working with
R. palustris because it was a very good model for studying anaerobic degradation of aromatic compounds but after sequencing its genome discovered that it was capable of doing more than what was known (Slonczewski,2009).The
R. palustris genome contains 5,459,213 base-pair circular chromosomes with 4,836 predicted genes and a plasmid of 8,427 bp. 1,514 genes, which is 31% of its genome, code for energy metabolism, biosynthesis, carbon & nitrogen metabolism and cellular processes.
R. palustris possess three nitrogenases, five benzene ring pathways and four light harvesting 2 systems which enable it to carry out its diverse metabolisms. It also has genes that enable it to oxidize hydrogen, thiosulfate and carbon monoxide and use them as energy sources.[2]
Degradation of Aromatic compounds
R. palustris is known to break down 3-chlorobenzoate via the reductive coenzyme A mediated pathway. 3-chlorobenzoate first gets converted to 3-chlorobenzoyl-CoA and then reductively dehalogenated to form benzoyl-CoA.Finally
R. palustris reduces benzoyl-CoA to form acetyl CoA and carbon dioxide. [4] The acetyl CoA can then enter the tricarboxylic acid cycle (TCA) and supply energy to the bacterium.In a study conducted,
R. palustris strain RCB 100 was grown on an anaerobic enrichment medium containing 2mM of 3-chlorobenzoate as the only source of organic source and incubated in light. The 3-chlorobenzoate gets dechlorinated first in the degradation and evidently chloride ions were produced in proportion to the amount of 3-chlorobenzoate consumed by the bacteria. The concentration of chloride ions was measured colorimetrically while the amount of 3-chlorobenzoate was determined by high pressure liquid chromatography (HPLC).
Hydrogen Production
R.palustris produces hydrogen as a byproduct of nitrogen fixation and it is possible to tap the hydrogen that is produced for human uses. Hydrogen is a far better fuel compared to coal or petroleum because the combustion of hydrogen only generates water.
R.palustris is unique compared to the other purple bacteria in that it codes for three different nitrogenases. It encodes a vanadium encoding nitrogenase that catalyzes the production of hydrogen three times as much as the common molybdenum-containing nitrogenases do.[1]
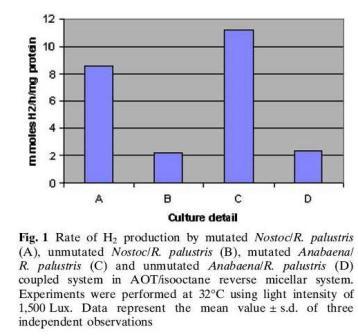
R.palustris has been coupled with other microbes such as cyanobacteria to form microreactors for hydrogen production. Cyanobacteria is also capable of nitrogen fixation. Cyanobacteria has three enzymes involved in hydrogen metabolism namely nitrogenase, membrane bound hydrogenase and soluble hydrogenase. Since the hydrogenases consume hydrogen in their course of action, mutants were produced by knocking off the gene encoding hydrogenase to maximize the production of hydrogen. The cyanobacteria mutants paired with
R.palustris had a 4.8 fold higher hydrogen production than the unmutated cyanobacteria/
R.palustris pair, 11.8mmol H2/h/mg protein vs. 3.9 mmol H2/h/mg protein, shown in Figure 1. The reactions were monitored within reversed micelles made of N-ethyl hexyl sodium sulfosuccinate. [3]
Nanoparticle synthesis
Living organisms have innate abilities to precisely direct the shape and crystallinity of inorganic materials, their abilities can be taken advantage of in the synthesis of inorganic materials. Likewise
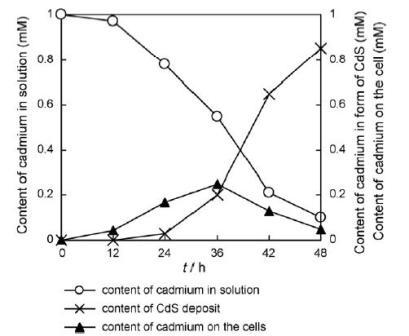
R.palustris is capable of synthesisizing cadmium sulfide (CdS) nanoparticles. When R. palustris was incubated in a solution of cadmium sulfate, the color of solution changed to yellow after 48 hours indicating the formation of cadmium sulfide nanoparticles. The
R.palustris cytoplasm contains cysteine disulfhydrase, an enzyme which catalyses the production of S2-, was responsible for the formation of CdS nanoparticles. R.palustris also secreted proteins that then stabilized the nanoparticles.
R.palustris produced stable and uniform nanoparticles of an average size of 8.01±0.25nm, which was confirmed by TEM analysis.
Conclusion
Overall text length at least 3,000 words, with at least 3 figures.
References
[1]"
Rhodopseudomonas palustris CGA009" Joint Genome Project Institute
[4][http://www.ncbi.nlm.nih.gov/pmc/articles/PMC92743/ Egland,P.G.,Gibson,J.,Harwood,C.,"Reductive, Coenzyme A-Mediated Pathway for 3-Chlorobenzoate Degradation in the Phototrophic Bacterium
Rhodopseudomonas palustris". "Applied and Environmental Microbiology".2001.Volume 67. p.1396-1399
Edited by student of Joan Slonczewski for BIOL 238 Microbiology, 2010, Kenyon College.