Snodgrassella alvi wkB2
Classification
Taxonomy
Domain: Bacteria
- Phylum: Proteobacteria
- Class: Betaproteobacteria
- Order: Neisseriales
- Family: Neisseriaceae
- Genus: Snodgrassella
- Species: alvi
- Strain: wkB2
- Strain: wkB2
- Species: alvi
- Genus: Snodgrassella
- Family: Neisseriaceae
- Order: Neisseriales
- Class: Betaproteobacteria
|
NCBI: Taxonomy |
Names
Snodgrassella alvi wkB2
Candidatus Snodgrassella alvi wkB2 (previously)
S. alvi wkB2
Description and Significance
S. alvi wkB2 is a core member of the western honey bee ( Apis mellifera ) worker gut microbiota [7]. The cells are short gram negative rods. On agar, S. alvi wkB2 forms smooth white round colonies [4]. It is a is non-motile biofilm former colonizing the ileum of A. mellifera [4].
Snodgrassella was so named in honor of Robert Evans Snodgrass who was a prominent figure studying insect physiology and had described Apis mellifera [6]. alvi refers to the latin word "alvus" which refers to the bowels or to bees [6]. The strain was described by Waldon Kwong who's initials are "w.k."
Initially, the name "Candidatus" was added to the front of Snodgrassella alvi because its discovery was through gene sequencing and had not been cultivated yet [6].
Genome Structure
Genome
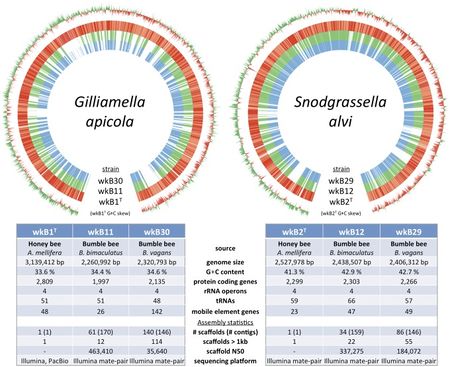
The genome of S. alvi wkB2 consists of a singular circular chromosome containing 2,527,978 bp [3]. The G+C content is 41.3% [3].
Some pieces of the genome have mutated over time and lost function as S. alvi wkB2 is an obligate symbiont of A. mellifera and is believed to have co-evolved [3]. S. alvi wkB2 is also beleived to engage in Horizontal Gene Transfer (HGT) with a proximal gut inhabitant of A. mellifera, Gilliamella apicola .
Annotated Genes
There are 2295 annotated genes and 2193 of them are protein coding, 76 are non-coding, the rest (26) are pseudogenes [8].
A few notable annotated gene functions include genes that code for many pathogen associated functions. One gene codes for nonribosomal peptide synthesis of siderophores and one that codes for type 6 secretion system (T6SS) [3].
Cell Structure, Metabolism and Life Cycle
Cell Structure
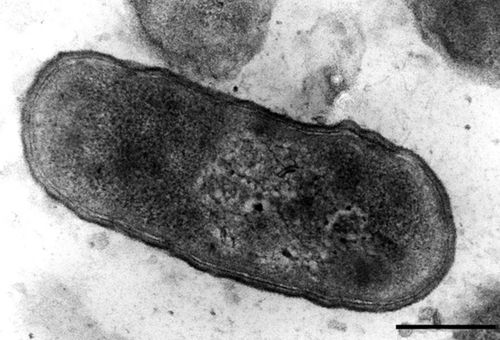
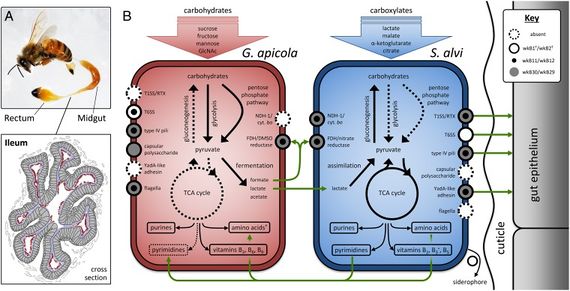
S. alvi wkB2 is gram negative, non-motile rod [4]. It displays a type IV pilli structure and YadA-like adhesins which are used to interact with the host gut epithelium which can be seen in figure 3. A TEM of S. alvi wkB2 can be seen in figure 1.
Metabolism
S. alvi wkB2 is an obligate aerobe. S. alvi wkB2 has transportation systems for taking up carboxylates like citrate, malate, & α-ketoclutarate [3]. These can then be used in the TAC cycle [3]. Transportation systems within S. alvi wkB2 can also allow for the uptake of lactate which can be directly converted into pyruvate through the enzyme, lactate dehydrogenase [3]. A TCA cycle enzyme, succinyl-CoA synthetase, which catalyzes the interconversion of succinyl-CoA and succinate, is not present in S. alvi yet the pathway is considered to still be functional as no alternative pathways exist to bypass the TCA cycle [3].
S. alvi is able to synthesize purine and pyrimidine nucleotides as well as all 21 proteinogenic amino acids (except selenocysteine). Vitamin B2 and B6 can be synthesized. S. alvi wkB2 specifically can synthesize B3 and its derivatives (NAD+ and NADP+) from aspartate [3].
Lost Metabolic Pathways
Evidence has been found suggesting that S. alvi was able to take up and break down carbohydrates, but has since lost that ability [3]. The Glycolosis pathway, Pentose phosphate pathway, & Entner-Doudoroff pathway have all lost essential enzymes [3]. These pathways are typically used to convert sugars to pyruvate.
Biochemical Tests
Nitrate reductase: Positive
Catalase: Positive
Urease: Positive
Cytochrome C Oxidase: Negative
β-Glucosidase: Negative
Indole production: Negative
Gelatinase: Negative
β-Galactosidase: Negative
Arginine dihydrolase: Negative
H2S production: Negative
Haemolysis: Negative
The above data was from the initial cultivation-based characterization of S. alvi wkB2 [4].
Life Cycle
S. alvi wkB2 lives its entire natural life within the gut of A. mellifera worker bees. Cultivation of S. alvi wkB2 in vivo needs no substrate or assistance outside of the bee's regular diet. It is primarily passed on through the fecal-oral route but can also be passed on from surfaces or bee socialization [6].
This strain can also be cultivated in vitro on the following agars: Heart Infusion, Heart Infusion supplemented with Sheep's Blood, Columbia supplemented with Sheep's Blood (CBA), & R2 agar [1]. Liquid cultivation of S. alvi wkB2 is typically on Insectargro, Salt-free LB, or R2 Liquid Media [1].
On solid or liquid media, in vitro cultivation of S. alvi wkB2 is typically at 35°C and 5% CO2 [1].
Ecology
Bee Gut Importance
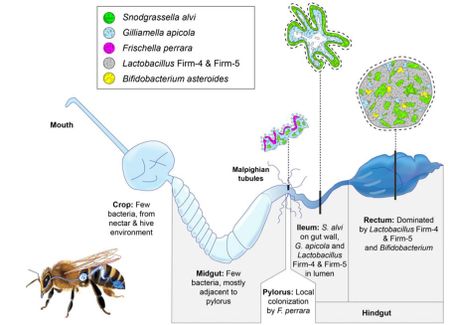
Within the bee gut, S. alvi wkB2 plays a vital role in O2 consumption keeping the gut of the bee anaerobic [5]. S. alvi wkB2 is also a primary biofilm former, forming a layer around the ileum which can be seen in figure 4 [5].
S. alvi wkB2 is able to occupy a very important niche within the bee gut, using carboxylates rather than carbohydrates suggesting that it has synergistic interactions with other native gut dwelling microbes which secrete this carboxylates [3]. However, S. alvi wkB2 is able to colonize and thrive within A. mellifera on its own.
Ecological Importance
For A. mellifera two pests that have been attributed to the large decline in honey bee populations are, the Varroa mite (Varroa destructor) and the Deformed Wing Virus (DWV) [2].
A transformed S. alvi wkB2 cells using RNAi have been proposed as a solution to the decreasing honey bee population [2]. It has been shown that these synthetic S. alvi wkB2 cells can sucessfully colonize A. mellifera in the same part of the ileum that it usually occupies [2][5].
This engineered microbe was transformed with a plasmid coding for double stranded RNA (dsRNA) targeting an essential mRNA on the deformed wing virus which plagues the western honey bee [2]. Leonard et al. were able to show an increase in bee survival for bees incoulated with the DWV and had sucessful colonizations of transformed S. alvi wkB2 cells [2].In another experiment, S. alvi wkB2 was transformed with a plasmid which coded for dsRNA which targeted 14 essential Varroa mite genes. They were also able to show a decrease in survival rates for the Varroa mites feeding on these bees [2].
Synthetically engineered S. alvi wkB2 after further testing and ethical consideration may be able to be implemented in the environment as a solution to protect the decreasing bee population.
References
[1] Barricklab.org. 2020. Barrick Lab :: Protocolsculturingsnodgrassellaalvi. [online] Available at: <https://barricklab.org/twiki/bin/view/Lab/ProtocolsCulturingSnodgrassellaAlvi> [Accessed 29 April 2020].
[2] Leonard, S., Powell, J., Perutka, J., Geng, P., Heckmann, L., Horak, R., Davies, B., Ellington, A., Barrick, J. and Moran, N., 2020. Engineered symbionts activate honey bee immunity and limit pathogens. Science, 367(6477), pp.573-576.
[3] Kwong, W., Engel, P., Koch, H. and Moran, N., 2014. Genomics and host specialization of honey bee and bumble bee gut symbionts. Proceedings of the National Academy of Sciences, 111(31), pp.11509-11514.
[4] Kwong, W. and Moran, N., 2012. Cultivation and characterization of the gut symbionts of honey bees and bumble bees: description of Snodgrassella alvi gen. nov., sp. nov., a member of the family Neisseriaceae of the Betaproteobacteria, and Gilliamella apicola gen. nov., sp. nov., a member of Orbaceae fam. nov., Orbales ord. nov., a sister taxon to the order 'Enterobacteriales' of the Gammaproteobacteria. INTERNATIONAL JOURNAL OF SYSTEMATIC AND EVOLUTIONARY MICROBIOLOGY, 63(Pt 6), pp.2008-2018.
[5] Kwong, W. and Moran, N., 2016. Gut microbial communities of social bees. Nature Reviews Microbiology, 14(6), pp.374-384.
[6] Martinson, V., Moy, J. and Moran, N., 2012. Establishment of Characteristic Gut Bacteria during Development of the Honeybee Worker. Applied and Environmental Microbiology, 78(8), pp.2830-2840.
[7] Moran, N., Hansen, A., Powell, J. and Sabree, Z., 2012. Distinctive Gut Microbiota of Honey Bees Assessed Using Deep Sampling from Individual Worker Bees. PLoS ONE, 7(4), p.e36393.
[8] Ncbi.nlm.nih.gov. 2020. Home - Gene - NCBI. [online] Available at: <https://www.ncbi.nlm.nih.gov/gene> [Accessed 29 April 2020].
Author
Page authored by Samantha Worthington, student of Prof. Jay Lennon at Indiana University.