Sulfolobus solfataricus: Difference between revisions
| Line 24: | Line 24: | ||
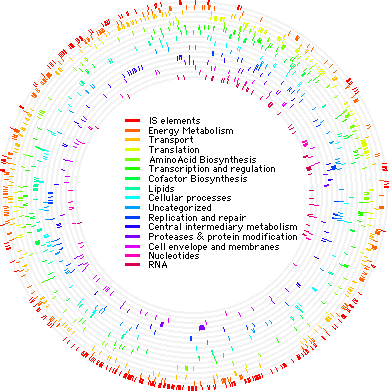
[[Image:Chrom.GIF|frame|left|Chromosome illustration of ''S. solfataricus'' showing gene function and location. Image courtesy of Qunxin She ''et al.'' (2001).]] | [[Image:Chrom.GIF|frame|left|Chromosome illustration of ''S. solfataricus'' showing gene function and location. Image courtesy of Qunxin She ''et al.'' (2001).]] | ||
The genome of ''S. solfataricus'' P2 was sequenced in 2001. It consists of a single circular chromosome containing 2,992,245 base pairs. There are 3,032 genes which encode 2,977 proteins. It has been shown that approximately one-third of the ''S. solfataricus'' P2 genome is unique. However, much of the genome (≈40%) is archaeal, while small portions overlap with bacteria and eukarya (12 and 2.3%, respectively). It has its own 743 open reading frames, and about 11% of the genome is mobile elements. | The genome of ''S. solfataricus'' P2 was sequenced in 2001. It consists of a single circular chromosome containing 2,992,245 base pairs. There are 3,032 genes which encode 2,977 proteins. It has been shown that approximately one-third of the ''S. solfataricus'' P2 genome is unique. However, much of the genome (≈40%) is archaeal, while small portions overlap with bacteria and eukarya (12 and 2.3%, respectively). It has its own 743 open reading frames, and about 11% of the genome is mobile elements. | ||
Revision as of 22:44, 15 April 2009
Classification
Archaea; Crenarchaeota; Thermoprotei; Sulfolobales; Sulfolobaceae
Species
|
NCBI: Taxonomy |
Sulfolobus solfataricus
Description and Significance
Sulfolobus solfataricus grow in volcanic hot springs where they have ample sulfur and low pH. S. solfatricus is a very widely studied crenarchaeal organism. For this reason it is used as a model organism in archaeal research, with DNA replication, the cell cycle, chromosomal integration, and RNA processing is well researched and understood.
Proteins found in S. solfataricus are being investigated for use in biotechnology because they are active in a wide range of temperatures.
Genome Structure
The genome of S. solfataricus P2 was sequenced in 2001. It consists of a single circular chromosome containing 2,992,245 base pairs. There are 3,032 genes which encode 2,977 proteins. It has been shown that approximately one-third of the S. solfataricus P2 genome is unique. However, much of the genome (≈40%) is archaeal, while small portions overlap with bacteria and eukarya (12 and 2.3%, respectively). It has its own 743 open reading frames, and about 11% of the genome is mobile elements.
The S. solfataricus P2 genome has already been used extensively. It has been integral in the study of crenarchaeal biology, and has been compared to many euryarchaea to elucidate differences between the two groups. It has also provided more evidence for the separations of archaea, eukarya, and bacteria. The S. solfataricus P2 genome has also been manipulated for DNA studies of transcription, translation, the cell cycle, and other genetic events. Currently, the Sulfolobus solfataricus 98/2 strain is being sequenced for further studies.
Cell Structure, Metabolism and Life Cycle
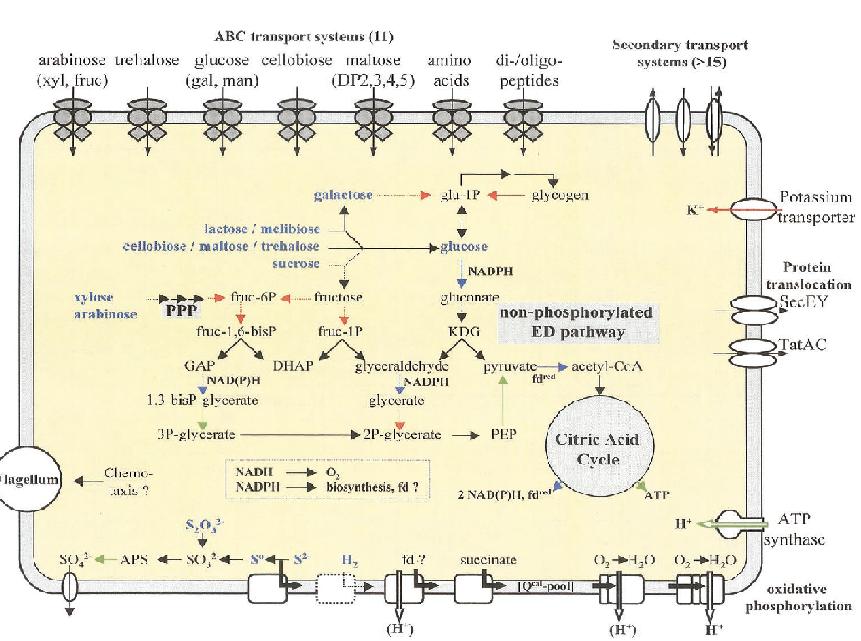
Sulfolobus solfataricus is an archaeal bacteria with a spherical shape and a clockwise flagellum. Its outer wall is unusual in that it does not contain any peptidoglycan. The cell wall contains - proteins that mediate the transport of essential carbohydrates into the cell; ion channels that keep potassium levels at vital concentrations; ATPase that uptakes protons from oxidative phosphorylation; and proton pumps that are vital for oxidative phosphorylation.
S. solfataricus utilizes the glycolitic pathway to convert glucose to pyruvate that is used in the citric acid cycle. Glucose is not available in the environment, and so s. solfataricus converts the carbohydrates present into glucose at an expense of energy. Glycolysis yields no net energy, however the citric acid cycle produces some ATP and reduces NADP+ to NADPH that can be used to drive oxidative phosphorylation. S. solfataricus also utilizes the pentose phosphate pathway when as a method of obtaining NADPH from carbohydrates that do not go through the glycolitic pathway.
S. solfataricus is able to synthesize all 20 amino acids.
When necessary, s. solfataricus can utilize sulfide and hydrogen as electron donors. As it's final electron acceptor s. solfataricus requires oxygen.
Ecology and Pathogenesis
Sulfolobus solfataricus is a thermoacidophile, and was first characterized in sulfur-rich volcanic springs in Yellowstone National Park. Strains of S. solfataricus have also been found in hot mud pools in the Solfatara crater north of Naples, Italy. It has also been discovered in El Salvador and the Dominica. While the optimum temperature for growth is ≈75oC, S. solfataricus can live in a range of 55-90oC. It can live in a pH range of 0.9-5.8, with its optimum being 2-3. However, S. solfataricus maintains its cytoplasmic pH at 6.5. It has also been shown that S. solfataricus requires an aerobic environment, in which it exists as a free-living sulfur chemolithotroph.
Strains of S. solfataricus are known to harbor plasmids that encourage bacteria-like conjugation between cells. However, the genes behind these mechanisms are dissimilar between S. solfataricus and bacteria.
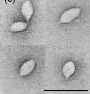
S. solfataricus can be infected by morphologically unusual viruses, such as SSV1 (Sulfolobus spindle-shaped virus). The virions formed by SSV1 often cluster together, forming rosette-like shapes. These viruses have been found in Yellowstone National Park, Iceland, and Japan.
References
Author
Page authored by Alvin Makohon-Moore and Antoni Malachowski, students of Prof. Jay Lennon at Michigan State University.