The CRISPR Immune System in Bacteria and Archaea: Difference between revisions
No edit summary |
No edit summary |
||
| Line 16: | Line 16: | ||
==Structure of CRISPR Loci== | ==Structure of CRISPR Loci== | ||
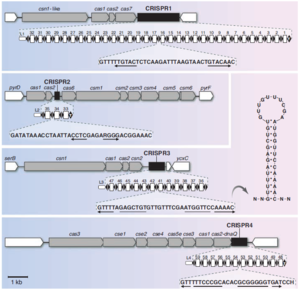
<br>CRISPR loci consist of alternating repeat sequences and spacer sequences (Figure 1). The repeat sequences (typically between 23 and 47 bp) are at least partially palindromic | <br>CRISPR loci consist of alternating repeat sequences and spacer sequences (Figure 1). The repeat sequences (typically between 23 and 47 bp) are at least partially palindromic. This means that the two halves of the sequence are complementary, allowing them to base pair. Because of this, after these sequences are transcribed, they are able to form hairpin shaped secondary structures. Repeat sequences are usually highly conserved within a CRISPR locus, but can vary greatly between different loci. The spacer sequences (typically 21-72 bp) originate from fragments of captured foreign DNA. Spacer segments may correspond to sequences of coding or non-coding DNA or RNA from viruses, phages, plasmids, or transposons. However, they most commonly target phage DNA. CRISPR loci are usually found in the cell’s genome, but can occur on plasmids [http://www.sciencemag.org/content/327/5962/167 (Horvath and Barrangou. 2010)]. | ||
CRISPR sequences encode the information necessary to identify unwanted foreign DNA. However, in order to defend the cell, CRISPR sequences require a collection of CRISPR associated proteins, called cas proteins. These proteins add new spacers to CRISPR. They also use RNA transcripts of existing spacers to identify and destroy phage DNA. These cas genes are usually adjacent to CRISPR sequences, often in the form of operons. There are a huge variety of cas genes associated with different CRISPR sequences. However, six core cas genes, designated cas1 through cas6, have been identified. All CRISPR/cas systems include these genes, as well as at least one of a set of subtype-specific genes [http://www.sciencemag.org/content/327/5962/167 (Horvath and Barrangou. 2010)]. | CRISPR sequences encode the information necessary to identify unwanted foreign DNA. However, in order to defend the cell, CRISPR sequences require a collection of CRISPR associated proteins, called cas proteins. These proteins add new spacers to CRISPR. They also use RNA transcripts of existing spacers to identify and destroy phage DNA. These cas genes are usually adjacent to CRISPR sequences, often in the form of operons. There are a huge variety of cas genes associated with different CRISPR sequences. However, six core cas genes, designated cas1 through cas6, have been identified. All CRISPR/cas systems include these genes, as well as at least one of a set of subtype-specific genes [http://www.sciencemag.org/content/327/5962/167 (Horvath and Barrangou. 2010)]. | ||
Revision as of 19:18, 9 May 2013
by Hunter Gervelis
Introduction
CRISPR (clustered regularly interspaced palindromic repeats) is a defense mechanism, present in bacteria and archaea, which confers immunity against phages. All species of bacteria and archaea are parasitized by viruses known as phages. Accordingly, prokaryotes have evolved many different types of protection against infection by phages and other sources of foreign DNA. These include prevention of absorption, blocking of injection, abortive infection, and the restriction-modification system (Horvath and Barrangou. 2010). CRISPR DNA sequences and their associated proteins are one such type of protection. The CRISPR system protects prokaryotic cells by destroying viral DNA after it has entered the cell.
Phages infect prokaryotic cells by binding to surface proteins, injecting their DNA through the cell wall, and hijacking the cell’s protein machinery to replicate the DNA. If, for any reason, the DNA is destroyed before it can cause infection, small fragments (21-72 bp) are integrated into dedicated loci (called CRISPR loci) within the cell’s genome. Later, if the cell encounters foreign DNA, it will compare it to the short stored sequences. If the sequences of the stored and foreign DNA match, the foreign DNA will be destroyed by enzymes (Horvath and Barrangou. 2010).
The CRISPR system’s ability to precisely and reliably cleave DNA has made it an active area of study for purposes of genetic engineering (Jinek et al., 2013).
Structure of CRISPR Loci
CRISPR loci consist of alternating repeat sequences and spacer sequences (Figure 1). The repeat sequences (typically between 23 and 47 bp) are at least partially palindromic. This means that the two halves of the sequence are complementary, allowing them to base pair. Because of this, after these sequences are transcribed, they are able to form hairpin shaped secondary structures. Repeat sequences are usually highly conserved within a CRISPR locus, but can vary greatly between different loci. The spacer sequences (typically 21-72 bp) originate from fragments of captured foreign DNA. Spacer segments may correspond to sequences of coding or non-coding DNA or RNA from viruses, phages, plasmids, or transposons. However, they most commonly target phage DNA. CRISPR loci are usually found in the cell’s genome, but can occur on plasmids (Horvath and Barrangou. 2010).
CRISPR sequences encode the information necessary to identify unwanted foreign DNA. However, in order to defend the cell, CRISPR sequences require a collection of CRISPR associated proteins, called cas proteins. These proteins add new spacers to CRISPR. They also use RNA transcripts of existing spacers to identify and destroy phage DNA. These cas genes are usually adjacent to CRISPR sequences, often in the form of operons. There are a huge variety of cas genes associated with different CRISPR sequences. However, six core cas genes, designated cas1 through cas6, have been identified. All CRISPR/cas systems include these genes, as well as at least one of a set of subtype-specific genes (Horvath and Barrangou. 2010).
CRISPR loci typically consist of fewer than 50 repeat-spacer alternations, although some include several hundred. Genomes may include one or several different CRISPR loci. In extreme cases, these loci can constitute over 1% of the genome. CRISPR loci are found in roughly 40% of bacterial genomes and 90% of archaea genomes. It is currently not known why they are more common in archaea. CRISPR sequences can be transmitted both horizontally and vertically (Horvath and Barrangou. 2010).
History
CRISPR sequences were first discovered in 1987 in Escherichia coli. They were observed in other bacterial species and in archaea in 2002. It was first suggested that CRISPR sequences are part of an immune system in 2005. This was due to the discovery of homology between spacer sequences and viral and plasmid DNA. Much has been learned about the function of cas proteins by disabling cas genes. For example, disabling the cas7 gene disrupts the cell’s ability to incorporate new spacers. Disabling the csn1-like gene causes a loss of resistance against phages, even if the relevant spacers are present. This shows that cas genes are necessary for immunity in addition to CRISPR sequences (Horvath and Barrangou. 2010).
Operation of the CRISPR/cas System
In general, operation of the CRISPR/cas system takes place in four steps. First, spacers corresponding to fragments of DNA from a phage or another source are incorporated into CRISPR. Little is known about this process. However, it is believed to involve the Cas1 and Cas2 proteins. Also, new spacers are added at the leading end of the CRISPR sequence. Second, CRISPR loci must be transcribed into pre-crRNA. A noncoding sequence at the leading end of CRISPR, rich in adenine and thymine, acts as a promoter for this purpose. Third, pre-crRNA must then be processed into crRNA. Each piece of crRNA consists of a single spacer between two half-repeats. Finally, after processing, the crRNA is used to neutralize foreign genetic material (Horvath and Barrangou. 2010).
The CRISPR/cas system is similar in principle to the RNA interference used by eukaryotic cells; both use short RNA sequences to guide the destruction of foreign DNA by enzymes. However, these two systems employ entirely different sets of proteins. No homology has been found between CRISPR/cas and RNAi (Horvath and Barrangou. 2010), (Lintner et al., 2011).
Although the alternating spacer repeat structure of CRISPR sequences is conserved across all species, the protein machinery which uses crRNA to destroy foreign genetic material is diverse. In some cases, highly intricate complexes composed of many different proteins are used to destroy foreign DNA (Wiedenheft et al., 2011). In other cases, a single protein with a guide RNA may be sufficient to cleave DNA (Cong et al., 2013). Three important examples of these protein mechanisms are CASCADE, the CMR complex, and Cas9. The CRISPR-associated complex for antiviral defense (cascade) degrades foreign DNA. The CMR complex uses crRNA to neutralize foreign RNA. Cas9 nuclease can accurately cleave DNA, and has great promise for genetic engineering.
CRISPR-associated Complex for Antiviral Defense (CASCADE) Targets Phage DNA
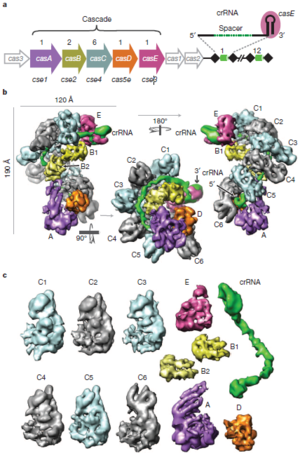
CASCADE is a protein-RNA complex which binds to foreign genetic material. If it binds strongly to foreign DNA, its conformation changes. This change marks the DNA for destruction by the enzyme Cas3. CASCADE targets single or double stranded DNA for destruction, but can bind complementary RNA as well. The structure of CASCADE has been mapped in great detail using cryo-electron microscopy (Figure 2). In E. coli, CASCADE consists of 11 protein subunits, including five different proteins, and one piece of crRNA (consisting of one pathogen-derived spacer between two half repeats). These proteins are designated CasA through CasE. Each CASCADE complex includes 1 copy each of CasA, CasD, and CasE, two copies of CasB, and six copies of CasC. Overall, the CASCADE has a “seahorse-like” shape. The six CasC subunits form a helical strand. This strand is the backbone of the complex. Pieces of crRNA fit into the groove of this helical strand. In this configuration, the crRNA can base pair with foreign DNA, but is protected from degradation by enzymes. CasE is at the “head” of the seahorse. It is the endonuclease that processes pre-crRNA into crRNA. It identifies pre-crRNA by binding the loop structures formed by the palindromic repeats. It then cleaves a piece of crRNA from the pre-crRNA. CasA and CasD form the “tail” of the seahorse. It has been shown that CasA plays a role in distinguishing self from foreign DNA. The two copies of CasB form a long dimer. This dimer lies along the CasC helix, connecting the head of the seahorse (CasE) to the tail (CasA) (Wiedenheft et al., 2011).
Some regions of the crRNA molecule are more important than others. Nucleotides 1-5 and 7-8 at the 5’ end of the spacer portion of the crRNa are known as the seed sequence. The 5’ end of the crRNA appears to be most important for binding foreign DNA targets. It has been shown that binding between DNA and crRNA is strongest at the 5’ end, and becomes weaker near the 3’ end of the crRNA. Also, phages with point mutations in DNA corresponding to the seed sequence evade detection by the CRISPR/cas system. Base pairing between crRNA and DNA begins at the seed sequence, and then continues towards the 3’ end of the crRNA. However, the crRNA and DNA do not base pair along their entire lengths. Rather, they form duplexes only four or five base pairs long. The crRNA binds to CasC subunits between these duplexes. Base pairing shortens the crRNA molecule, causing the entire CASCADE complex to change shape. It has been shown that CasE, CasB, and CasA all change orientation during base pairing. This conformational change signals the bound DNA for destruction by Cas3 (Wiedenheft et al., 2011).
The CASCADE complex varies between different species of bacteria and archaea. However, some of its core elements are conserved. For example, a CASCADE complex in sulfolobus Solfataricus contains proteins analogous to CasC and CasD in E. coli (Lintner et al., 2011).
The CMR Complex Targets Phage RNA
The CMR (cas RAMP module complex, where RAMP stands for repeat associated mysterious protein) complex is a protein-RNA complex which uses crRNA to destroy foreign RNA (usually mRNA transcribed from phage DNA). It is also known as the type IIIB CRISPR/cas system. Like CASCADE, this complex can be found in both bacteria and archaea. However, its structure and mechanism of action are very different from CASCADE. The CMR complex both identifies and destroys RNA. This differs from CASCADE, which instead identifies suspicious DNA and triggers its destruction by other enzymes. It also lacks CASCADE’s characteristic “seahorse” structure. Instead, it is shaped like a “crab claw”. Because the CMR complex targets RNA and not DNA, there is no need for a PAM sequence. PAM sequences are only needed to distinguish foreign DNA from the cell genome. As with CASCADE, the CMR complex shows great variability across species (Zhang et al., 2011).
Adaptations by Phages
Prokaryotes and phages are locked in an ongoing evolutionary arms race. Whenever a new defense mechanism emerges in bacteria, phages eventually mutate to circumvent that defense. The CRISPR/cas system is no exception. In response to the emergence of the CRISPR/cas system, some phages have acquired mutations which allow them to infect “immunized” cells. Mutations with this effect include point mutations within the proto-spacer. The proto-spacer is the segment of the phage genome which corresponds to the CRISPR spacer. Immunity may also be lost if there is a mutation to the proto-spacer adjacent motif (PAM). PAMs are short (2-3 bp), highly conserved sequences adjacent to proto-spacers. They may be required to distinguish foreign DNA from its own genome (Horvath and Barrangou. 2010).
More recently, anti-CRISPR genes have been discovered in bacteriophages, which can allow phages to successfully infect bacteria with CRISPR. This is true even if the cell contains a spacer specific to the invading phage. The precise mechanism by which these genes subvert CRISPR/cas is not known. However, it has been shown that they do not hamper transcription of cas proteins or crRNA. There is also evidence that they do not target particular spacers. These genes are not broadly effective against all CRISPR/cas systems. A particular set of anti-phage genes permits a phage to infect only cells with a particular class of CRISPR/cas systems (Bondy-Denomy et al. 2013).
Classification of CRISPR/cas Systems
The nomenclature of cas genes and proteins can be confusing at times. Different sources may provide different names for cas proteins or for subtypes of CRISPR/cas systems. This is due to disagreement over the homology of the proteins, and how they should be classified into families. However, recent progress has been made in developing an agreed upon classification system. CRISPR/cas systems have been classified into three groups based on which genes they include. These groups are designated types I, II, and III. The only two genes common to all three categories are cas1 and cas2, both of which are believed to be involved in the acquisition of new spacers. Each type of CRISPR system is defined by the presence of a signature gene. For type I, II, and III systems, these genes are cas3, cas9, and, cas10, respectively. Each type can be further subdivided based on the presence or absence of other genes (Makarova et al., 2011).
CRISPR and Antibiotic Resistance
Interestingly, it has been shown that the CRISPR/cas system may slow the spread of antibiotic resistance genes. In addition to phage DNA, the CRISPR/cas system can destroy plasmids. Horizontal transfer of plasmids containing resistance genes is a common mechanism for the spread of antibiotic resistance. If the CRISPR/cas system targets plasmids containing resistance genes, transformation will not take place. This has been tested by transfecting two strains of S. epidermidis with two different plasmids. One strain contained a CRISPR locus with a spacer targeting a nickase gene, the other had no CRISPR locus (nickase is an enzyme which cleaves only one strand of a double stranded DNA molecule). While one plasmid included the naturally occurring nickase gene, the other plasmid contained a form of the gene with several silent mutations. These silent mutations permitted the gene to function, but prevented it from matching the CRISPR spacer. The strain of S. epidermidis lacking a CRISPR locus accepted both plasmids via conjugation from S. aureus. The strain with a CRISPR locus accepted only the plasmid with the mutant form of the gene. This is of interest because the nickase gene is very common on staphylococci plasmids. It may be feasible to manipulate the CRISPR/cas system to slow the spread of antibiotic resistance genes in a clinical setting (Marraffini et al., 2008).
Cas9 and Genetic Engineering Applications of CRISPR
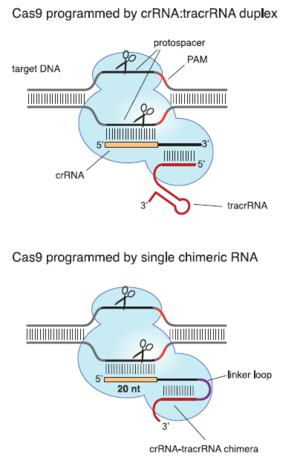
Type II CRISPR/cas systems can be used to precisely and reliably cleave double stranded DNA. This has made them a topic of great interest for genetic engineering. Type II systems are of particular interest partially because of their simplicity compared to the multi-protein CASCADE complex. In Eukaryotic cells, just the Cas9 endonuclease and its guide RNA are sufficient to cleave target DNA (Cong et al., 2013). Like all CRISPR/cas systems, type II systems use crRNA to identify and silence foreign genetic material. However, unlike type I or type III systems, they also require a piece of trans-activating RNA (tracrRNA). These tracrRNAs are short sequences that are partially complementary to the corresponding crRNA. They are required for Cas9 to recognize foreign DNA (Figure 3). It is possible that they correctly orient the crRNA so that it can bind to foreign DNA. These RNAs form a complex with the Cas9 protein, which then cleaves foreign DNA. Instead of a crRNA-tracrRNA pair, a single piece of chimeric RNA can be designed and used to activate Cas9 (Jinek et al., 2013). The chimeric strand can fold on itself via complementary base pairing. The resulting hairpin structure mimics the shape of the crRNA-tracrRNA sufficiently to allow Cas9 to cleave DNA. However, in some cases this design is less effective than the naturally occurring crRNA-tracrRNA duplex (Jinek et al., 2013). This opens the possibility of using type II CRISPR/cas systems to edit genomes by selectively and accurately cleaving DNA (Cong et al., 2013).
The type II CRISPR/cas system can even be used to edit the genomes of human cells. Specifically, it has been shown that proteins and RNA from the CRISPR/cas system of Streptococcus pyogenes can selectively introduce double stranded breaks into human chromosomes. This is done by transfecting human cells with DNA encoding the required cas genes, a CRISPR sequence targeting the desired loci within the genome, and other necessary elements. However, successful editing requires the system to be modified from its naturally occurring form in prokaryotes. In Streptococcus pyogenes, the CRISPR locus includes four genes. However, only the nuclease Cas9 and the requisite tracrRNA and crRNA are necessary for introducing double stranded breaks in human DNA. In Streptococcus pyogenes and other prokaryotes, host factor ribonuclease III (RNase III) is also necessary for DNA cleavage. However, mammalian cells apparently contain their own nucleases which fulfill its role in crRNA processing. The sequences of Cas9 and RNase III used for human genome editing are codon optimized. That is, for each amino acid, the codon which permits the fastest and most accurate translation in human cells is selected. In addition to the cas genes and CRISPR sequence, a nuclear localization signal must be included. This tags the CRISPR/cas system to be taken into the nucleus. Finally, suitable promoters must be included for both the cas genes and CRISPR sequence. When all of these elements are present, efficient and accurate DNA cleavage is possible in human and other mammalian cells. Its effectiveness is comparable to or better than other editing methods, such as TALE nucleases. Cas9 is highly site specific. A single base pair mismatch between crRNA and the target loci can prevent cleavage (Cong et al., 2013). Furthermore, this system is highly versatile. By including multiple spacers targeting different genome loci, it is possible to edit the genome in many places at once. Two concurrent double stranded breaks can be used to delete targeted sequences. However, the CRISPR/cas system still has some limitations. It can only cleave DNA at loci where there is a suitable PAM sequence. Although naturally occurring Cas9 cleaves double stranded DNA, it is possible to mutate the protein so that it becomes a nickase (cleaves only one DNA strand). In the future, it may be possible to fully disable the endonuclease activity of Cas9, so that it selectively binds DNA loci without cleavage (DiCarlo et al., 2013).
It has also been shown that Cas9 can be used to help make cells integrate donor DNA into their genomes. Cas9 and its guide RNA are used to introduce double stranded breaks at the loci where the new DNA is to be added. The donor DNA can then be integrated into the cell’s genome when the break is repaired. When Cas9 cleaves double stranded DNA in vivo, the break is repaired by homologous recombination. The repair process often results in mutations. These mutations are useful for research, as the mutation rate can be used to measure the efficacy of DNA cleavage. Furthermore, these mutations make it possible to use Cas9 to knock out genes. This has been successfully demonstrated by introducing oligonucleotides at selected loci in yeast genomes. The Cas9 gene, RNA targeting the desired loci, and the necessary promoters and nuclear localization signals where introduced using a plasmid. This technique may make it possible to introduce donor DNA into a cell’s genome with high specificity and a high recombination rate compared to other methods (DiCarlo et al., 2013).
Conclusion
Infection by phages has forced bacteria and archaea to evolve a diverse arsenal of defense mechanisms against these viruses. CRISPR sequences and their associated proteins are a particularly sophisticated example of such a defense mechanism. The CRISPR/cas system stores phage DNA sequences. Later, cas proteins use the stored sequences to identify and destroy phage DNA before the cell can be destroyed. Sequences corresponding to phage DNA or RNA are incorporated as spacers into dedicated CRISPR loci within the cell’s genome. These sequences are then transcribed to produce short segments of crRNA. The crRNA is used by cas proteins to bind and destroy foreign DNA. The details of this process vary, and cas proteins are highly diverse. Recently, the CRISPR/cas system has emerged as a powerful and useful genetic engineering tool. It is desirable for the ease with which it can be programmed to cleave specific DNA sequences and for its high accuracy. CRISPR/cas and its uses remain an exciting area for future research.
References
1. [Bondy-Denomy, Joe. et al. 2013. Bacteriophage Genes that Inactivate the CRISPR/Cas Bacterial Immune System. Nature 493: 429-434.] http://au8dt3yy7l.search.serialssolutions.com/?ctx_ver=Z39.88-2004&ctx_enc=info%3Aofi%2Fenc%3AUTF-8&rfr_id=info:sid/summon.serialssolutions.com&rft_val_fmt=info:ofi/fmt:kev:mtx:journal&rft.genre=article&rft.atitle=Bacteriophage+genes+that+inactivate+the+CRISPR%2FCas+bacterial+immune+system&rft.jtitle=Nature&rft.au=Bondy-Denomy%2C+Joe&rft.au=Pawluk%2C+April&rft.au=Maxwell%2C+Karen+L&rft.au=Davidson%2C+Alan+R&rft.date=2013-01-17&rft.eissn=1476-4687&rft.volume=493&rft.issue=7432&rft.spage=429&rft_id=info:pmid/23242138&rft.externalDocID=23242138
2. [Cong, Le. et al. 2013. Multiplex Genome Engineering Using CRISPR/cas Systems. Science 339: 819-823.] http://www.sciencemag.org/content/339/6121/819
3. [DiCarlo, James E. et al. 2013. Genome Engineering in Saccharomyces cerevisiae using CRISPR/Cas Systems. Nucleic Acids Research 41(7): 4336-4343.] http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3627607/?tool=pmcentrez&rendertype=abstract
4. [Philippe Horvath and Rodolphe Barrangou 2010. CRISPR/Cas, the Immune System of Bacteria and ARchaea. Science 327: 167-170.] http://www.sciencemag.org/content/327/5962/167
5. [Jinek, Martin. et al. 2013. A Programmable Dual RNA Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 337: 816-821.] http://www.sciencemag.org/content/337/6096/816
6. [Lintner, Nathanael G. et al. 2011. Structural and Functional Characterization of an Archaeal Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR)-ASSOCIATED Complex for Antiviral Defense (CASCADE). Journal of Biological Chemistry 286: 21643-21656.] http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3122221/?tool=pmcentrez&rendertype=abstract
7. [Makarova, Kira S. et al. 2011. Unification of Cas Protein Families and a Simple Scenario for the Origin of CRISPR/cas Systems. Biology Direct 6.] http://www.biology-direct.com/content/6/1/38
8. [Marraffini, Luciano A. and Sontheimer, Erik J. 2008. CRISPR Interference Limits Horizontal Gene Transfer in Staphylococci by Targeting DNA. Science 322: 1843-1845.] http://www.sciencemag.org/content/322/5909/1843
9. [Wiedenheft, Blake. et al. 2011. Structures of the RNA-Guided Surveillance Complex from a Bacterial Immune System. Nature 477: 486-490.] http://www.nature.com/nature/journal/v477/n7365/full/nature10402.html
10. [Zhang, Jing. et al. 2011. Structure and Mechanism of the CMR Complex for CRISPR-Mediated Antiviral Immunity. Molecular Cell 45: 303-313.] http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3381847/?tool=pmcentrez&rendertype=abstract
Edited by student of Joan Slonczewski for BIOL 238 Microbiology, 2009, Kenyon College.
