The Implications of Broadly Neutralizing Antibody Development and Epitope Targeting for HIV Vaccine Development: Difference between revisions
| Line 2: | Line 2: | ||
==Introduction== | ==Introduction== | ||
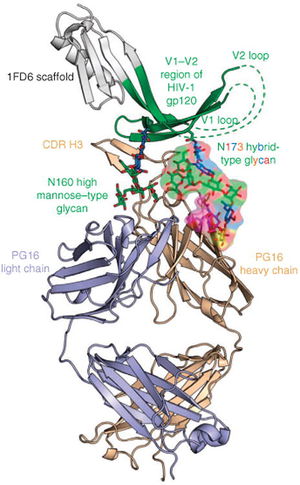
[[Image:Example HIV Antibody.jpeg|thumb|300px|right|Figure 1. Pancera M. et al. Structural basis for diverse N-glycan recognition by HIV-1-neutralizing V1-V2 directed antibody PG16. The light chain of the broadly neutralizing antibody is represented in light purple and the heavy chain is represented in tan. This figure highlights two sections of the glycan shield that are vital to antibody binding of the HIV spike. The glycan N160 is represented in green and the glycan N173 represented as many colors to represent its hybrid nature. Both of these glycans are a part of the V1-V2 region of the gp120, an HIV envelope spike protein. The CDR (Complementary determining region) of the antibody is shown making direct contact with these two vital spikes. <i> Nature Structural & Molecular Biology</i> 2013. | [[Image:Example HIV Antibody.jpeg|thumb|300px|right|Figure 1. Pancera M. et al. Structural basis for diverse N-glycan recognition by HIV-1-neutralizing V1-V2 directed antibody PG16. The light chain of the broadly neutralizing antibody is represented in light purple and the heavy chain is represented in tan. This figure highlights two sections of the glycan shield that are vital to antibody binding of the HIV spike. The glycan N160 is represented in green and the glycan N173 represented as many colors to represent its hybrid nature. Both of these glycans are a part of the V1-V2 region of the gp120, an HIV envelope spike protein. The CDR (Complementary determining region) of the antibody is shown making direct contact with these two vital spikes. <i> Nature Structural & Molecular Biology</i> 2013. Image Source : http://www.nature.com/nsmb/journal/v20/n7/full/nsmb.2600.html | ||
) ]] | ) ]] | ||
Revision as of 04:59, 17 December 2014
By Sean Smith
Introduction
Robert Gallo and Luc Montagnier discovered Human immunodeficiency virus to be the causative agent of Autoimmune deficiency syndrome (AIDS) in 1984 only a few years after the initial HIV outbreak across the world. [1] However, unlike other viruses known to cause epidemics across the world, such as polio, no effective vaccine for HIV has been discovered through classical strategies. HIV’s high rate of mutation, greater than any other characterized human pathogen [2], is thought to be responsible for the repeated failure of attempts to develop a vaccine. Due to the high rate of mutations, the target sites of the antibodies, the epitopes, are constantly changing. Furthermore, an antibody with specific binding to a single epitope may lose its binding affinity in response to mutations in a target epitope. The decrease in binding affinity eliminates the neutralizing ability of the antibody and allows viral progression to proceed unmitigated.
However, in a select percentage of chronically infected HIV patients, potent antibodies known as broadly neutralizing antibodies have been shown to effectively bind a wide range of distinct epitopes. The increased range of binding affinity allows a single clade of similar broadly neutralizing antibodies to neutralize many distinct mutations of HIV. The characteristics of broadly neutralizing antibodies have attracted the attention of vaccine researchers who believe that a vaccine that causes the development of potent broadly neutralizing antibodies could effectively prevent early HIV infection. Research to better understand the development of HIV antibodies and the structures of their epitopes has given weight to the belief that broadly neutralizing antibodies have the potential to effectively prevent early HIV progression. The following is a summary of the current research on antibody epitopes and antibody development and of the further characterization needed to effectively translate such research into an effective vaccine.
Structures of Primary HIV Antibody Epitopes
All primary HIV antibody epitopes are found on the heterotrimer HIV envelope spike (Figure 1). The heterotrimer spike is comprised of gp41, a protein partially embedded in the HIV membrane that binds other envelope proteins [3], and gp120, the protein responsible for binding CCR5, CD4, or CXCR4 [4]. A complex glycan layer surrounds gp41 and gp120 to comprise much of the outer layer of the HIV spike [5]. The range of epitopes is greatly varied. Antibodies have been found to target specific segments of the gp41 protein and the gp120 protein in both glycan shield dependent and independent manners and have been shown to depend on the quaternary structure of the spike in some instances while showing structure independent binding in other cases.
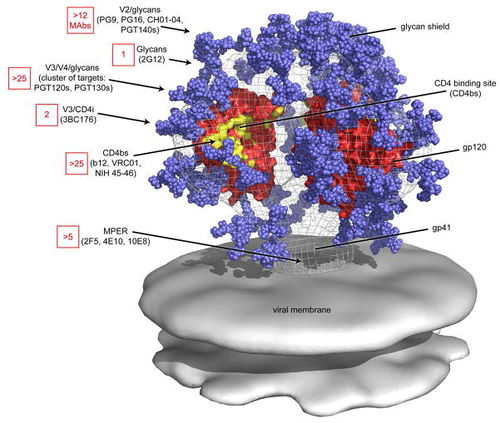
Four antibody epitopes on the HIV spike that have been shown to be targets for broadly neutralizing antibodies are MPER, the CD4 binding site, the V1/V2 loop and the V3 loop [6]. The following is a summary of the known structural characteristics and of the antibodies known to target such sites. MPER (Membrane proximal external region) is a highly conserved target for neutralizing antibodies located on the gp41 protein in the base of the HIV spike [7]. The broadly neutralizing neutralizing antibody 10E8 has been shown to bind to highly conserved hydrophobic residues of the MPER with specificity [8]. The CD4 binding site on the gp120 spike protein is targeted by broadly neutralizing antibodies that mimic CD4 binding. The broadly neutralizing antibody VCR01 interacts with the CD4 binding site and causes a conformational shift at the binding site through its interaction [9]. The conformational shift depresses the functionality of the receptor and therefore neutralizes the HIV virus.
The V1/V2 (Variable region 1/Variable Region 2) loop on the gp120 protein is one of the most frequently mutated portions of the HIV virion. However, the V1/V2 loop has been shown to be one of the most frequent epitopes for potent broadly neutralizing antibodies [10]. Known antibodies PG9 and VCR26 appear to rely on the conservation of quaternary structure and on the conversation of the N linked glycan at residue 160 of the V1/V2 loop heighten antibody/epitope binding affinity [11]. The effects of the potential mutations at that site will be discussed below. The V3 (Variable region 3) loop on the gp120 plays a vital role in viral entry into the host cell [12]. The exposed tip of the V3 loop has been targeted by neutralizing in a quaternary-structure specific manner. However, many V3 loop targeting antibodies have narrow, rather than broad, neutralizing specificity. More research on how the structure of V3 governs antibody specificity is needed [5].
The glycan shield surrounding gp120 and gp41 on the HIV spike plays a vital role in determining many antibody epitopes. N-linked glycans determine almost half of the mass of the HIV spike. However, very few glycan dependent or glycan specific neutralizing antibodies have been isolated and epitopes appear to be centralized on the gp120 protein and to a lesser extent at the base of the gp41 protein [5]. Despite the fact that antibodies have not been shown to bind to the glycan shield directly, the shield can still affect antibody binding through steric hindrance. Since the glycan shield covers over half of the entire spike, it governs the how accessible specific potential epitopes are to neutralizing antibodies [13].
Common Mutations in the Structure of Primary Epitopes
Due to the incredible rate of mutation in the HIV genome, even some of the highly conserved regions of the HIV spike undergo shifts that induce conformational and amino acid shifts. Many of these changes can alter the ability of a neutralizing antibody to both access and bind a potential epitope. The conformational and sequential shifts most deleterious to antibody binding ability are known as escape mutations. Escape mutations at a specific site cause a neutralizing antibody to lose the ability to effectively bind at that site and lead to a loss of that antibody's ability to mitigate HIV viral progression [14]. The following is a summary of some of characterized escape mutations at vital binding sites. A better understanding of escape mechanisms will aid in understanding how many broadly neutralizing antibodies develop to effectively neutralize epitopes with differential structural and amino acid components in response to HIV spike mutations [15].
Alterations in the conformation of the glycan shield of the HIV spike can constitute an effective escape mutation [14]. Often, changes in the glycan shield conformation do not decrease the the binding affinity between the neutralization antibody and its target epitope. Instead, such glycan shield changes obstructs the antibody from reaching its target site through steric hindrance [14]. Therefore, the size and shape of an antibody may serve as a vital factor in determining an antibody's ability to continue to effectively neutralize an HIV virion in the face of shifts in the glycan shield.
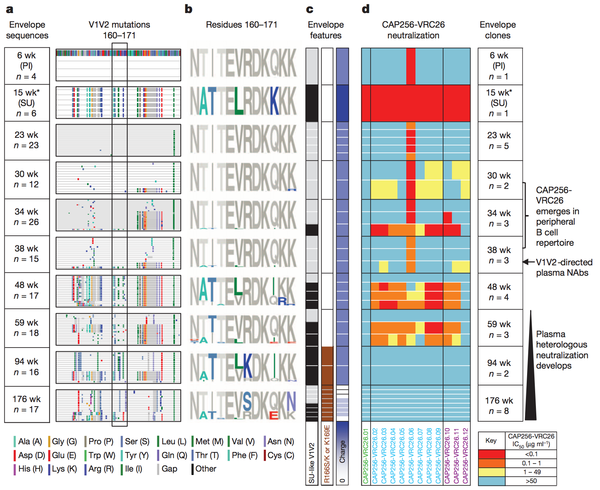
Amino acid shifts at key residues in target epitopes can decrease the binding affinity between the neutralizing antibody and the target epitope. Doria-Rose et al., characterized such an escape mutation at residues 169 and 166 of the V1V2 sequence (Figure 3) The presence of the escape mutation shifts R166S/K and K169E is shown by the brown squares in column 2 of panel c of the figure. Such mutations were categorized as escape mutations because the drastically nullified the ability of existing neutralizing antibodies to effectively prevent HIV viral production. As noted in panel 4 of figure 3 the ability of the antibodies to neutralize HIV virions present at weeks 94 and 176 after the foundation of the escape mutation at week 59 plummeted.
Interestingly, and not surprisingly, the advent of this escape mutation altered antibody development in patient CAP256 studied by Doria-Rose et al. The antibodies produced after the initial escape mutation decreased in neutralizing specificity and rapidly increased in neutralizing breadth [16]. Therefore, researcher now hypothesize that escape mutations may be a vital factor driving the natural production of broadly neutralizing antibodies. The ability to harness such a hypothesis to artificially guide broadly neutralizing antibody development is discussed below.
HIV Antibody Development in a Single Patient
The invention of next generation sequencing has opened the door towards the understanding of many molecular level phenomenon. One phenomenon that can now be extensively traced is the development of neutralizing antibodies in HIV over time. Through sequence similarity analysis of the heavy and light chains of a line of somatically related antibodies, one can trace how the lineage of antibodies changed from its unmutated common ancestor over time [17]. HIV antibodies change in response to the ever mutating HIV virus. Therefore, tracing antibody change over time sheds light the development of the HIV virus and on the rate of change in antibody development. Characterization of both phenomenon is essential to assessing the validity of broadly neutralizing antibodies as a potential vaccine component for if broadly neutralizing antibodies never underwent rapid development, their potential as a vaccine response would decrease.
Fortunately, thorough sequence analysis reveals not only that antibodies develop rapidly in response to changes the HIV virus population but also that a massive range of distinct but somatically related antibodies develop from a common ancestor. Furthermore, the somatically related antibodies have drastically different neutralizing potentials [16] and the difference in neutralization potential is dependent on the week post-infection from which the antibody was isolated [18]. Such findings confirmed the hypothesis that neutralization potential changes of a single lineage of antibodies changes over time.
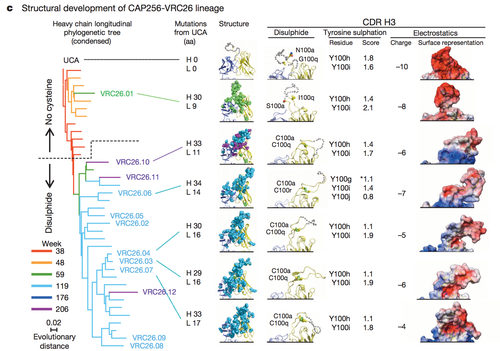
Figure 4 highlights some of the structural differences that arise as a result of somatic mutation in a single antibody lineage and how such structural difference lead to differential antibody epitope recognition on a molecular level [16]. The complementary determining region 3 loop is a section of the antibody VRC26 integral in determining epitope binding. Three factors were examined to assess the structural differences in the CDR H3 loops: disulphide bonding, tyrosine sulfation and electrostatic potential. In this patient CAP256 studied by Doria-Rose, a vital escape mutation presented itself at week 59. Figure 4 demonstrates the structural evidence of altered antibody development as a result of the escape mutation.
The surface representation of the electrostatic potential of antibody VRC26.01 greatly resembles the electrostatic potential of the unmutated common ancestor (UCA) shown in row one. Not surprisingly, the structural representations of the electrostatic potential of the antibodies isolated after week 59 differ from the surface representations of the UCA and VCR26.01 but resemble each other. A similar trend can be noted in the analysis of tyrosine sulfation and disulphide bond formation [16].
The characterization of the complex events that drive the production of diverse set of antibodies from a single lineage begins to elucidate just how complex the antibody response to HIV can be. Further characterization of the factors that drive the production of early specifically neutralization and of late broadly neutralizing is needed to fully understand how and if directed antibody development can be harnessed to yield and effective HIV vaccine.
Development of Broadly Neutralizing Antibodies and the Potential Role of Broadly Neutralizing Antibodies in Vaccine Development
Despite high levels of mutation in potential antibody binding sites on the HIV spike, 10 to 25% develop cross-reactive, or broadly, neutralizing antibodies after many years of exposure to HIV [13]. These broadly neutralizing antibodies have the ability to recognize and bind to a wide range or distinct epitopes as opposed to a single epitope on the HIV spike. Therefore, broadly neutralizing antibodies are more adept at effectively neutralizing the ever mutating HIV virus. Unfortunately, despite the great diversity of potential binding sites for these broadly neutralizing antibodies, the diversity of the HIV infection by the time of their genesis is too great to overcome, and broadly neutralizing antibodies in chronically infected patients often only has a minimal effect on HIV viral load and HIV disease progression.
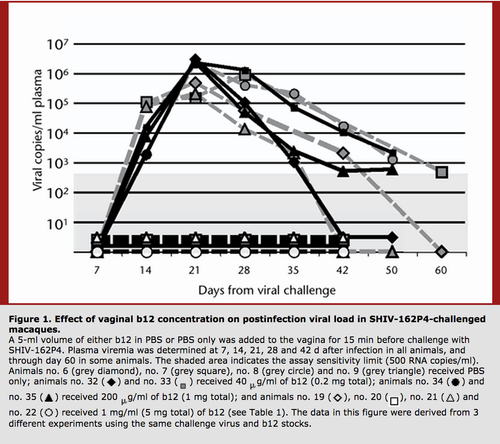
Interestingly, it is thought that broadly neutralizing antibodies would have a greater effect on decreasing HIV viral load in patients at the early stages of HIV infection. Furthermore treatment of macaques with the broadly neutralizing antibody b12 has been shown to protect from SHIV challenges. Therefore, while broadly neutralizing antibodies may not prevent the progression of chronic HIV, they can effectively prevent transmission when administered vaginally into a model organism [19]. Such findings increase the hopes that a vaccine that elicits the production of broadly neutralizing antibodies could prevent HIV transmission.
However, there has been no isolation of natural broadly neutralizing antibodies from sera of early stage HIV infected patients. No broadly neutralizing antibodies may exist in early stage infection due to the lower levels of mutations and quasi-species formation in the HIV virus and the sheer number of somatic mutations necessary to attain broad neutralization across a range of distinct epitopes [18]. On average, 70 total changes in the amino acid sequence occur during the development of a broadly neutralizing antibody from the unmutated ancestor [18]. For a vaccine to become feasible, a means to cause formation of broadly neutralizing antibodies in HIV negative patients would need to be discovered.
Role of T Follicular Helper Cells in Antibody Development:
T follicular helper (Tfh) cells have been shown to be integral in B-cell formation and differentiation at germinal centers, and general B-cell population management [4] The maintenance of standard B-cell population sizes aids the timely development of antibodies in response to shifts in the HIV population [20]. Therefore, any vaccine reliant on the development of broadly neutralizing antibodies will most likely have to contain a factor to ensure proper T follicular helper cell function.
While the precise mechanism by which Tfh cells regulate the differentiation of B cells at germinal centers, it is clear that proper Tfh function is required to ensure effective control of B cell differentiation with germinal centers [2]. Germinal centers are areas where mature B cells differentiate within secondary lymphoid tissue [21]. At the germinal center, the B cell can proceed to enter apoptosis, differentiate into a memory B cell, differentiate into a plasma cell, or maintain itself in the GC B cell state. The Tfh cell is central to determining which of these 4 processes occurs and is therefore critical in antibody development.
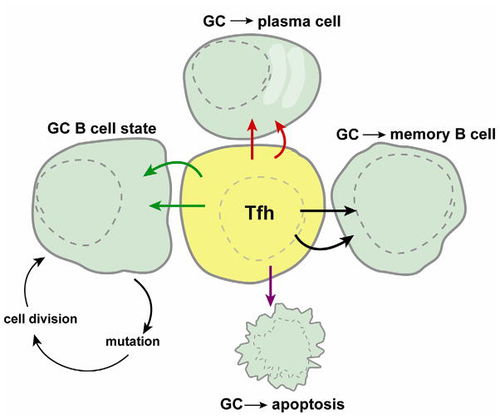
Tfh cells operate by a specific rather than a general mechanism and HIV specific Tfh cells can be isolated from the sera of HIV positive patients. In chronic infections, Tfh cells counts have been found to be highly elevated [20]. Under normal conditions, Tfh cell count is the limiting factor in B-cell differentiation, however, due to the high level of Tfh cells found during chronic HIV infection, this limit may no longer apply. Due to altered B-cell differentiation patterns as a result of elevated Tfh counts, Tfh cells may play a distinct role in the late infection alterations in immune response of HIV infections such as the development of broadly neutralizing antibodies.
Interestingly, high levels of affinity maturation, like those produced by elevated Tfh counts, have been linked to an increased breadth of neutralization ability of the antibodies produced through that process [22]. More research focused on Tfh regulatory mechanisms is needed. However, it is likely that control of Tfh function will play an important role in any HIV vaccine that may incorporate the production of broadly neutralizing antibodies
Future Research
While there is much evidence that an eventual HIV vaccine reliant on the production broadly neutralizing antibodies and Tfh cell regulation could eventually come into fruition, much research is needed before the medical dream could become a reality. More extensive characterization of the factors responsible for driving broadly neutralizing antibody production is needed. Currently, the immunogen responsible for signaling the production of broadly neutralizing antibodies has not been elucidated [4].
Furthermore, no means has been developed to quickly replicate the high levels of somatic mutation necessary to create broadly neutralizing antibodies. However, the evidence presented by Doria-Rose et al is encouraging since broadly neutralizing antibodies have been shown to develop quickly in response to changing stimuli [16].
In addition to further characterization of antibody development, further characterization of antibody binding sites and the HIV spike in general is needed. A catalog of common viable mutations to the HIV spike proteins and glycan shield would greatly aid the ability to predict mutation patterns in early HIV development and would give rise to a more effective vaccine design. While much research is needed, an effective HIV vaccine could come into being on the back of broadly neutralizing antibodies.
Conclusion
References
[1] "In Their Own Words: NIH Researchers Recall the Early Years of AIDS." NIH History. NIH. Web. 09 Nov. 2014.
[2] Sanjuan R. et al. Viral Mutation Rates Journal of Virology 2010.
[3] Stowell, Dan. "gp41" 2006. Web 09 Nov. 2014. http://www.mcld.co.uk/hiv/?q=gp41.
[4] Burton B.R. et al. A Blueprint For HIV Vaccine Discovery. Cell Host Microbe. 2012.
[5] Pancera M. et al. Structural basis for diverse N-glycan recognition by HIV-1-neutralizing V1-V2 directed antibody PG16. Nature Structural & Molecular Biology 2013.
[6] Hepler NL. et al. IDEPIL Rapid Prediction of HIV-1 Antibody Epitopes and Other Phenotypic Features from Sequence Data Using a Flexible Machine Learning Platform. PLOS Computational Biology 2014.
[7] Montero M. et al. The Membrane-Proximal External Region of the Human Immunodeficiency Virus Type 1 Envelope: Dominant Site of Antibody Neutralization and Target for Vaccine Design." Microbiology and Molecular Biology Reviews 2008.
[8] Huang J. et al. Broad and Potent Neutralization of HIV-1 by a gp41-Specific Human Antibody. Nature 2012.
[9] Scheid JF. et al. Sequence and Structural Convergence of Broad and Potent HIV Antibodies That Mimic CD4 Binding. Science 2011.
[10] Moore P.L. et al. Potent and Broad Neutralization of HIV-1 Subtype C by Plasma Antibodies Targeting a Quaternary Epitope Including Residues in the V2 Loop. Journal of Virology 2011.
[11] McLellan JS. et al. Structure of HIV-1 gp120 V1/V2 Domain with Broadly Neutralizing Antibody PG9. Nature 2012.
[12] Tian H. et al. Sequence Variation and Consensus Sequence of V3 Loo on HIV-1 gp120. Immunology Letters 2002.
[13] Ringe R., Bhattacharya J. Preventive and Therapeutics Applications of Neutralizing Antibodies to Human Immunodeficiency Virus Type 1 (HIV-1). Therapeutic Advances in Vaccines 2013.
[14] Wei X. et al. Antibody Neutralization and Escape by HIV-1. Nature 2003.
[15] Moore P.L. et al. Multiple Pathways of Escape from HIV broadly Cross-Neutralizing V2-Dependent Antibodies. Journal of Virology 2013.
[16] Doria-Rose NA. Developmental Pathway for Potent V1V2-Directed HIV Neutralizing Antibodies. Nature 2014.
[17] Zhu J. et al. Mining the Antibodyome for HIV-1-Neutralizing Antibodies with Next-Generation Sequencing and Phylogenetic Pairing of Heavy/Light Chains. PNAS 2013.
[18] Wu X. et al. Focused Evolution of HIV-1 Neutralizing Antibodies Revealed by Structures and Deep Sequencing. Science 2011.
[19] Veazey RS. et al. Prevention of Virus Transmission to Macaque Monkeys By a Vaginally Applied Monoclonal Antibody to HIV-1 gp120. Nature Medicine 2003.
[20] Lindqvist M. et al. Expansion of HIV Specific T Follicular Helper Cells in Chronic HIV Infection. The Journal of Clinical Investigation 2012.
[21] MacLennan IC. Germinal Centers. Annu Rev Immunol 1994.
[22] Zhou T. et al. Structural Basis for Broad and Potent Neutralization of HIV-1 by Antibody VRC01. Science 2010.
