Thermithiobacillus tepidarius: Difference between revisions
(→Genome) |
(→Genome) |
||
| Line 22: | Line 22: | ||
===Genome=== | ===Genome=== | ||
''T. tepdiarius'' [https://en.wikipedia.org/wiki/Genome genome] comprises of 2,968 predicted genes, within this strain 2,902 are protein-coding, 66 are RNA coding genes and 2 rRNA [https://en.wikipedia.org/wiki/Operon operons]. This particular strain genome size is [https://environmentalmicrobiome.biomedcentral.com/articles/10.1186/s40793-016-0188-0 2,958,498 bp-long]. There has been evidence of [https://en.wikipedia.org/wiki/Horizontal_gene_transfer horizontal gene transfer] which accounts for roughly 6% of the protein-coding genes found (Boden 2016).[[File:Thermithiobacullius genome.png|300px|thumb|left|[https://environmentalmicrobiome.biomedcentral.com/articles/10.1186/s40793-016-0188-0/tables/3 Figure 2: Permanant draft genome] of ''Thermithiobaciullis tepidarius'' DMS 3134 (Boden et. al., 2016).]] Including transfer from [https://en.wikipedia.org/wiki/Methylococcus_capsulatus ''Methylococcus capsulatus''] bath and ''Thiobacillus spp'', both isolated from the same spring in which it was found. While testing the strain, a [https://en.wikipedia.org/wiki/SOX_gene_family sox gene] cluster was found, very similar in its structure to those from other ''Acidithiobacillia''. An additional gene between soxA and soxB was found and annotated as a DUF302-family protein of unknown function. Since the [https://www.researchgate.net/figure/The-FriedrichKelly-model-for-the-Sox-mechanism-of-thiosulfate-oxidation-in_fig1_8633105 Kelly-Friedrich pathway] of thiosulfate oxidation is not used in multiple species of ''Thermithiobacillus'', the role of the operon (if there is any) remains unknown. Compared to other types of species, ''T. tepidarius'' DSM 3134 has one of the smallest genomes. Presumably, because ''T. tepidarius'' lacks the salt-tolerance system unlike species of ''Acidithiobacillia'' spp., ''Thiobacillus'' spp. and ''Halothiobacillus'' spp. Or even the iron-oxidation or acid-tolerance of multiple ''Acidithiobacillus'' species (Boden et. al., 2016). | ''T. tepdiarius'' [https://en.wikipedia.org/wiki/Genome genome] comprises of 2,968 predicted genes, within this strain 2,902 are protein-coding, 66 are RNA coding genes and 2 rRNA [https://en.wikipedia.org/wiki/Operon operons]. This particular strain genome size is [https://environmentalmicrobiome.biomedcentral.com/articles/10.1186/s40793-016-0188-0 2,958,498 bp-long]. There has been evidence of [https://en.wikipedia.org/wiki/Horizontal_gene_transfer horizontal gene transfer] which accounts for roughly 6% of the protein-coding genes found (Boden et. al.,2016).[[File:Thermithiobacullius genome.png|300px|thumb|left|[https://environmentalmicrobiome.biomedcentral.com/articles/10.1186/s40793-016-0188-0/tables/3 Figure 2: Permanant draft genome] of ''Thermithiobaciullis tepidarius'' DMS 3134 (Boden et. al., 2016).]] Including transfer from [https://en.wikipedia.org/wiki/Methylococcus_capsulatus ''Methylococcus capsulatus''] bath and ''Thiobacillus spp'', both isolated from the same spring in which it was found. While testing the strain, a [https://en.wikipedia.org/wiki/SOX_gene_family sox gene] cluster was found, very similar in its structure to those from other ''Acidithiobacillia''. An additional gene between soxA and soxB was found and annotated as a DUF302-family protein of unknown function. Since the [https://www.researchgate.net/figure/The-FriedrichKelly-model-for-the-Sox-mechanism-of-thiosulfate-oxidation-in_fig1_8633105 Kelly-Friedrich pathway] of thiosulfate oxidation is not used in multiple species of ''Thermithiobacillus'', the role of the operon (if there is any) remains unknown. Compared to other types of species, ''T. tepidarius'' DSM 3134 has one of the smallest genomes. Presumably, because ''T. tepidarius'' lacks the salt-tolerance system unlike species of ''Acidithiobacillia'' spp., ''Thiobacillus'' spp. and ''Halothiobacillus'' spp. Or even the iron-oxidation or acid-tolerance of multiple ''Acidithiobacillus'' species (Boden et. al., 2016). | ||
===Structure, Metabolism=== | ===Structure, Metabolism=== | ||
Revision as of 17:20, 29 April 2020
Classification
Domain: Bacteria
Kingdom: Eubacteria
Phylum: Proteobacteria
Class: Acidithiobacillia
Order: Acidithiobacillales
Family: Thermithiobacillaceae
Genus: Thermithiobacillus
Species: Tepidarius
Description and Significance
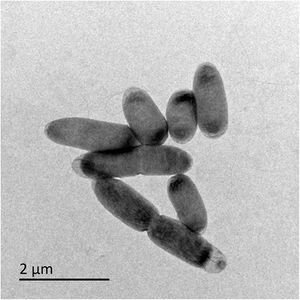
Thermithiobacillus tepidarius is a moderate thermophile and an obligate chemolithoautotrophy growing at the expense of reduced sulfur species (Berenguer et.al.,1970). T.tepidarius is also a non-spore-forming and gram-negative bacteria. This particular strain was isolated from sulfidic groundwater flowing into a Roman bathhouse in Bath, United Kingdom and the only other strain of this genus originated from decomposing concrete in the Melbourne sewers in the 1940s. In the past scientists have detected at least 6 OTUs representing other Thermithiobacillus species in 16S rRNA gene libraries from the Roman Baths, and there has been an isolated number of strains to date (Wood, Kelly et. al. 1986). This indicates that Thermithiobacillus species are as equally difficult to isolate as other sulfur-oxidizing autotrophs. Therefore, they may be simply rare or confined to ecosystems or environments that are rare.Thermithiobacillus tepidarius is significant because it's one of the only species of the genus with a published name. In addition, it's one of only two strains found in cultivation.
Genome
T. tepdiarius genome comprises of 2,968 predicted genes, within this strain 2,902 are protein-coding, 66 are RNA coding genes and 2 rRNA operons. This particular strain genome size is 2,958,498 bp-long. There has been evidence of horizontal gene transfer which accounts for roughly 6% of the protein-coding genes found (Boden et. al.,2016).
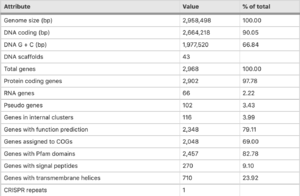
Including transfer from Methylococcus capsulatus bath and Thiobacillus spp, both isolated from the same spring in which it was found. While testing the strain, a sox gene cluster was found, very similar in its structure to those from other Acidithiobacillia. An additional gene between soxA and soxB was found and annotated as a DUF302-family protein of unknown function. Since the Kelly-Friedrich pathway of thiosulfate oxidation is not used in multiple species of Thermithiobacillus, the role of the operon (if there is any) remains unknown. Compared to other types of species, T. tepidarius DSM 3134 has one of the smallest genomes. Presumably, because T. tepidarius lacks the salt-tolerance system unlike species of Acidithiobacillia spp., Thiobacillus spp. and Halothiobacillus spp. Or even the iron-oxidation or acid-tolerance of multiple Acidithiobacillus species (Boden et. al., 2016).
Structure, Metabolism
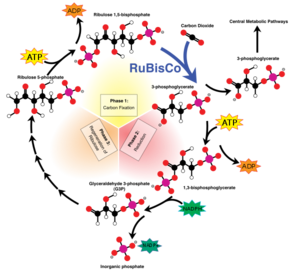
T. terpidarius has an optimum pH of 6.0-7.5. Even with this narrow range in pH, the rod shape cells can continue to grow on an acid medium of pH 4.8. However, they can only occur on reduced inorganic sulfur compounds and elementary sulfur. If grown on thiosulfate-containing basal salts agar, in forty-eight hours T. tepidarius forms white colonies of 2-5mm in diameter. These colonies smell faintly of elementary sulfur due to their intake of reduced sulfur species. Unlike other genera of Acidithiobacillus, T. tepidarius and multiple species of Thermithiobacillus are unable to oxidize iron-containing minerals or ferrous iron. This organism uses the carbon dioxide fixation pathway, biosynthesis, and sulfur oxidation as a way to obtain their nutrients. The cells fix carbon dioxide via the Calvin-Benson-Bassham cycle (Figure 3). The cells then gather polyphosphate ('volutin') granules as they are grown in batch cultures. These cultures are typically free from storage granules when they have grown in energy-source-limited chemostats. Their energy sources that support their autotrophic growth are elementary sulfur, sulfide tetrathionate, hexathionate, hepthathionate, and thiosulfate (Boden 2016).
Ecology and Known Roles in Symbiosis
This microbe will typically be found in environments with pH’s of 6.0-7.5 but can be found within unlikely ecosystems with pHs of 4.8 (Wood, Kelly et. al. 1986). This particular strain was found in a warm bath fed by natural thermal water, or the thermal groundwaters in Bath, United Kingdom. Other sulfur-oxidizing autotrophs very similar to T. tepidarius are prevalent in environments hosting both abundant reduced sulfur species and low oxygen concentrations. Environments such as marine sediments, OMZs or oxygen minimum zones, and hydrothermal systems. Based on this microbes genome sequence there will be future evolutionary studies into the nature of Acidithiobacilia and chemolithoautotrophs in general. In addition, the genome will further ecological studies including organism to organism interactions in the environment based on the evidence of the horizontal gene transfer found within.
References
Author
Page authored by Genevieve Grah, student of Prof. Jay Lennon at IndianaUniversity.
