Tobamovirus: Difference between revisions
No edit summary |
|||
| (10 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
{{Curated}} | |||
{{Viral Biorealm Genus}} | |||
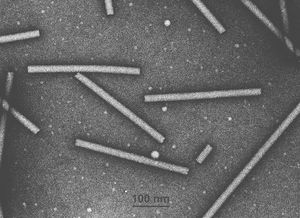
[[Image:tobamo1.jpg|thumb|right|EM of Tobacco Mosaic virus. From [http://www.ncbi.nlm.nih.gov/ICTVdb/Images/Milne/tobamo1.htm ICTVdb Picture Gallery], Courtesy Dr. RG Milne]] | |||
==Baltimore Classification== | ==Baltimore Classification== | ||
===Higher order taxa=== | ===Higher order taxa=== | ||
Viruses; ssRNA positive-strand viruses, no DNA stage; Tobamovirus | Viruses; ssRNA positive-strand viruses, no DNA stage; (no assigned family); ''Tobamovirus'' | ||
===Species=== | ===Species=== | ||
''Tobacco mosaic virus'', ''Cucumber green mottle mosaic virus'', ''Frangipani mosaic virus'', ''Turnip vein-clearing virus'' (examples) | |||
==Description and Significance== | ==Description and Significance== | ||
Tobamoviruses are one of the most studied plant viruses. Tobamoviruses are classified into two subgroups, defined by different genomic locations of their origin of virion assembly. (sources: Chen and Citovsky, [http://mbe.library.arizona.edu/data/1996/1310/4lart.pdf Lartey et al.]) | |||
==Genome Structure== | ==Genome Structure== | ||
The genome | The Tobamovirus genome is either monomeric or polymeric, non-segmented, and contains a single molecule of linear, positive-sense, single-stranded RNA. Minor species of non-genomic nucleic acid may also be found in virions. The encapsidated nucleic acid is mainly of genomic origin but virions may also contain nucleic acid of host origin and subgenomic mRNA including host rRNA found in the short particles of some species. The complete genome sequence is about 6450 nucleotides long. The 5'-end of the genome has a methylated nucleotide cap. The 3'-terminus has a tRNA-like structure that accepts histidine. (source: [http://www.ncbi.nlm.nih.gov/ICTVdb/ICTVdB/00.071.0.01.htm ICTVdB]) | ||
==Virion Structure of a Tobamovirus== | ==Virion Structure of a Tobamovirus== | ||
Tobamovirus virions consist of a non-enveloped capsid. The capsid is elongated, rod shaped, straight, and exhibits helical symmetry. Virons are 70-100 nm in length and 15-17.26-18.2 nm in width. The capsid has a herring-bone pattern with a low percentage in population clear predominate lengths. The axial canal could be distinct with a diameter of 2-3.257-4 nm. The basic helix can either be be obvious or obscure, and has a pitch of 2.3-2.314-2.4 nm. (source: [http://www.ncbi.nlm.nih.gov/ICTVdb/ICTVdB/00.071.0.01.htm ICTVdB]) | |||
==Reproduction Cycle of a Tobamovirus in a Host Cell== | ==Reproduction Cycle of a Tobamovirus in a Host Cell== | ||
==Viral Ecology & Pathology== | ==Viral Ecology & Pathology== | ||
Tobamoviruses are extremely stable. According to Castello et al., "For example, infectivity was reduced only 12-fold when purified tobacco mosaic tobamovirus (TMV) was exposed on the surface of a rocket to solar radiation for several minutes at an altitude of up to 149 km. The tobamoviruses survive over long periods of time under conditions favoring preservation." Tobamoviruses also have a wide host range, and have been detected in plants, soil, water, and clouds. There is no known insect vector, but it is known that tobamoviruses are transmitted through the sap of infected plants by plant to plant contact, handling by workers, on glasshouse structures and utensils, and by plant propagation. (source: Castello et al., Spence et al.) | |||
==References== | ==References== | ||
Castello et al. "Detection of tomato mosaic tobamovirus RNA in ancient glacial ice." ''Polar Biology'' 22.3 (1999): 207-212. | |||
Chen, Min-Huei and Vitaly Citovsky. "Systemic movement of a tobamovirus requires host cell pectin methylesterase." ''The Plant Journal'' 35.3 (2003): 386-392. | |||
[http://www.ncbi.nlm.nih.gov/ICTVdb/ICTVdB/00.071.0.01.htm ICTVdB - The Universal Virus Database, version 4. http://www.ncbi.nlm.nih.gov/ICTVdb/ICTVdB/] | |||
[http://mbe.library.arizona.edu/data/1996/1310/4lart.pdf Lartey et al. "Tobamovirus Evolution: Gene Overlaps, Recombination, and Taxonomic Implications." ''Molecular Biology and Evolution'' 13.10 (1996): 1327-1338.] | |||
Spence et al. "Characterisation of a Tobamovirus from Trailing Petunias." ''European Journal of Plant Pathology'' 107.6 (2001): 633-638. | |||
Latest revision as of 00:35, 8 August 2010
A Viral Biorealm page on the genus Tobamovirus
Baltimore Classification
Higher order taxa
Viruses; ssRNA positive-strand viruses, no DNA stage; (no assigned family); Tobamovirus
Species
Tobacco mosaic virus, Cucumber green mottle mosaic virus, Frangipani mosaic virus, Turnip vein-clearing virus (examples)
Description and Significance
Tobamoviruses are one of the most studied plant viruses. Tobamoviruses are classified into two subgroups, defined by different genomic locations of their origin of virion assembly. (sources: Chen and Citovsky, Lartey et al.)
Genome Structure
The Tobamovirus genome is either monomeric or polymeric, non-segmented, and contains a single molecule of linear, positive-sense, single-stranded RNA. Minor species of non-genomic nucleic acid may also be found in virions. The encapsidated nucleic acid is mainly of genomic origin but virions may also contain nucleic acid of host origin and subgenomic mRNA including host rRNA found in the short particles of some species. The complete genome sequence is about 6450 nucleotides long. The 5'-end of the genome has a methylated nucleotide cap. The 3'-terminus has a tRNA-like structure that accepts histidine. (source: ICTVdB)
Virion Structure of a Tobamovirus
Tobamovirus virions consist of a non-enveloped capsid. The capsid is elongated, rod shaped, straight, and exhibits helical symmetry. Virons are 70-100 nm in length and 15-17.26-18.2 nm in width. The capsid has a herring-bone pattern with a low percentage in population clear predominate lengths. The axial canal could be distinct with a diameter of 2-3.257-4 nm. The basic helix can either be be obvious or obscure, and has a pitch of 2.3-2.314-2.4 nm. (source: ICTVdB)
Reproduction Cycle of a Tobamovirus in a Host Cell
Viral Ecology & Pathology
Tobamoviruses are extremely stable. According to Castello et al., "For example, infectivity was reduced only 12-fold when purified tobacco mosaic tobamovirus (TMV) was exposed on the surface of a rocket to solar radiation for several minutes at an altitude of up to 149 km. The tobamoviruses survive over long periods of time under conditions favoring preservation." Tobamoviruses also have a wide host range, and have been detected in plants, soil, water, and clouds. There is no known insect vector, but it is known that tobamoviruses are transmitted through the sap of infected plants by plant to plant contact, handling by workers, on glasshouse structures and utensils, and by plant propagation. (source: Castello et al., Spence et al.)
References
Castello et al. "Detection of tomato mosaic tobamovirus RNA in ancient glacial ice." Polar Biology 22.3 (1999): 207-212.
Chen, Min-Huei and Vitaly Citovsky. "Systemic movement of a tobamovirus requires host cell pectin methylesterase." The Plant Journal 35.3 (2003): 386-392.
ICTVdB - The Universal Virus Database, version 4. http://www.ncbi.nlm.nih.gov/ICTVdb/ICTVdB/
Spence et al. "Characterisation of a Tobamovirus from Trailing Petunias." European Journal of Plant Pathology 107.6 (2001): 633-638.
