Transduction Pathways, Regulation and Significance of Iron Acquisition in Bacteria
The Importance of Iron in Bacteria
Iron is an element essential for bacterial growth. In its ferric form (Fe3+), iron can serve as a terminal electron acceptor (TEA) in an electron transport chain, being reduced to its ferrous form (Fe2+) [1]. Iron is also needed intracellularly as a cofactor or prosthetic group in cytochromes or in metabolic enzymes [2]. Thus, it is important for bacteria to be able to access iron. Iron exists in soluble ferrous form, or in ferric form where it is usually found as insoluble ferric oxides in the environment, or sequestered in host biological systems. In human hosts, iron can be found as heme, in hemoglobin, or bound by transferrin, an iron binding protein found in blood and tissues [2]. Therefore, high affinity methods for accessing iron have evolved in bacteria as a consequence of limited free ferric iron in the local environment.
High Affinity Siderophore Iron Acquisition
Energy Transduction for Siderophore Import

One high affinity iron chelating system that is well conserved in diverse bacteria involves secreted siderophores, which bind ferric iron with high affinity [3][4]. In Gram-negative bacteria, high specificity TonB-dependent outer membrane receptors are involved, allowing passage of the iron-siderophore complex into the periplasm using active transport [5]. The energy for this active transport is a consequence of TonB system energy transduction from the proton motive force (PMF).
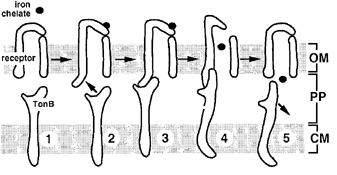
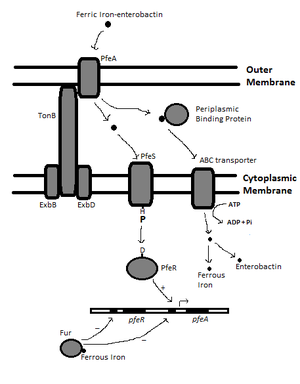
In Escherichia coli, the TonB system consists of TonB, ExbB, and ExbD, which are cytoplasmic membrane proteins [6]. These are coupled to FepA, the outer membrane receptor specific to enterobactin class siderophores (Fig. 1) [6]. Binding of the iron-siderophore complex to FepA induces a conformational change, exposing domains that cause TonB to interact with FepA (Fig. 2) [7]. This binding causes a conformational change in FepA using energy from the PMF derived from the cellular membrane, transduced through TonB (Fig. 2) [7]. This allows the iron-siderophore complex into the periplasm, and then into the cytoplasm via ATP-binding cassette (ABC) transporters (Fig. 3) [7]. Upon release of the ligand, FepA returns to its unstimulated state and TonB is released and readopts its high energy state from the PMF due to interactions with ExbB and ExbD [7]. Iron is released from siderophores through intracellular reduction to ferrous iron, as siderophores have lower affinity for ferrous iron (Fig. 3) [8].
Signal Transduction in TonB-dependent Receptors
The expression of TonB-dependent outer membrane receptors is induced through positive feedback mediated by a signal transduction pathway involving a two-component system [9]. In Pseudomonas aeruginosa, PfeA is an enterobactin siderophore receptor similar to FepA in E. coli [9]. PfeA expression is induced by periplasmic iron-siderophore complex binding to PfeS, a sensor kinase located at the cellular membrane (Fig. 3) [9]. Substrate binding induces a conformational change in PfeS, allowing autophosphorylation of a specific histidine residue [9]. Once phosphorylated, PfeS-P can in turn phosphorylate the response regulator, PfeR, at a key asparagine residue (Fig. 3) [9]. The activated PfeR-P can then bind to the pfeA promoter to induce the transcription of PfeA (Fig. 3) [9]. Thus, the presence of iron-siderophore complexes in the extracellular environment increases the expression of PfeA for more efficient uptake of iron from the environment.
Regulation of Iron Acquisition
As much as bacteria need iron to undergo essential cellular processes, iron uptake must be regulated. Excessive intracellular iron can lead to hydroxyl radical formation through the interaction between ferrous iron and hydrogen peroxide in the Fenton reaction [3]. In Gram-negative bacteria, regulation occurs through a ferric uptake regulator (Fur) regulon, which uses a conserved Fur protein [10]. Once bound to intracellular ferrous iron, Fur serves as a transcriptional repressor for genes encoding siderophores, TonB, outer membrane receptors, and other gene products related to iron acquisition (Fig. 3) [10]. Therefore, once a certain concentration of iron has been achieved in the cytoplasm of the bacterium, iron uptake genes will be downregulated, ensuring that an optimal concentration of iron is maintained in the cell.
Impact and Significance of Iron Acquisition
Significance on Virulence
In pathogenic bacteria, one of the key requirements for virulence is the ability of the bacteria to access iron. Not only does iron serve as an essential element for many biological processes, but access to iron can also alter the expression of key virulence factors [2]. For example, in humans, iron is complexed as heme, sequestered from siderophores in erythrocytes (Fig. 4) [2]. Hemolysin, a secreted protein that lyses erythrocytes to release heme for bacterial acquisition, is a key virulence factor in pathogenic bacteria such as Escherichia spp., Pseudomonas spp., Shigella spp., and Vibrio spp., regulated by Fur at the transcriptional level [11][12]. Also, in the absence of free iron, these pathogenic bacteria have been demonstrated to lower the pH in the host tissues to encourage pH-dependent dissociation of iron bound to transferrin [12]. Thus, it can be inferred that some key virulence factors are needed to disrupt host processes to obtain iron for use in the bacteria.
It has also been shown that when the free iron concentration in the host increases, bacterial virulence increases [[[#References|[12]]]]. This may be the result of the increasing bioavailability of iron for the incorporation into essential enzymes necessary to cause infection. In E. coli and P. aeruginosa, the expression of factors that influence iron acquisition are well described and provide models for bacterial virulence induction. This may allow the development of pharmaceuticals that target the iron acquisition systems of pathogenic bacteria to aid in the treatment of infection.
Ecological and Environmental Significance
In aerobic soil environments, plants have been shown to exhibit mutualistic relationships with microorganisms in order to acquire iron. It is critical that the soil is aerobic as bacteria require oxygen for siderophore synthesis [13]. Some plants contain microbial siderophore receptors as well as their own siderophore-like compounds, utilizing high affinity iron chelation systems to obtain iron for intracellular use [13]. In some iron deficient soil environments, microorganisms and plant microbe mutualists compete for free iron through siderophore production [13]. This promotes the evolution of versatile siderophores and receptors that have increasingly high affinity and specificity [13]. Thus, the microbial populations that secrete siderophores are important in controlling iron turnover in the soil through acquisition into biological systems followed by release through organic matter decay [13].
References
[1] Glasauer, S., Langley, S. and Beveridge, J.T. (2002). “Intracellular Iron Minerals in a Dissimilatory Iron-Reducing Bacterium.” Science, 295(5552): 117-119. DOI: 10.1126/science.1066577
[2] Martinez, J.L., Delgado-Ilibarren, A., and Baquero, F. (1990). “Mechanisms of iron acquisition and bacterial virulence.” FEMS Microbiology Reviews, 75(1): 45-56. DOI: 10.1016/0378-1097(90)90522-R
[3] Bagg, A. and Neilands, J.B. (1987). “Molecular Mechanism of Regulation of Siderophore-Mediated Iron Assimilation.” Microbiological Reviews, 51(4): 509-518.
[4] Miethke, M. and Marahiel, M.A. (2007). “Siderophore-Based Iron Acquisition and Pathogen Control.” Microbiology and Molecular Biology Reviews, 71(3): 413-451. DOI: 10.1128/MMBR.00012-07
[5] Ferguson, A.D. and Deisenhofer, J. (2002). “TonB-dependent receptors–structural perspectives.” Biochimica et Biophyisica Acta – Biomembranes, 1565(2): 318-332. DOI: 10.1016/S0005-2736(02)00578-3
[6] Higgs, P.I., Larsen, R.A. and Postle, K. (2002). “Quantification of known components of the Escherichia coli TonB energy transduction system: TonB, ExbB, ExbD and FepA.” Molecular Microbiology, 44(1): 271-281. DOI: 10.1046/j.1365-2958.2002.02880.x
[7] Moeck, G.S. and Coulton, J.W. (1998). “TonB-dependent iron acquisition: mechanisms of siderophore-mediated active transport.” Molecular Microbiology, 28(4): 675-681. DOI: 10.1046/j.1365-2958.1998.00817.x
[8] Andrew, S.C., Robinson, A.K. and Rodriguez-Quinones, F. (2003). “Bacterial Iron Homeostasis.” FEMS Microbiology Reviews, 27(2): 215-237. DOI: 10.1016/S0168-6445(03)00055-X
[9] Crosa, J.H. (1997). “Signal Transduction and Transcriptional and Posttranscriptional Control of Iron-Regulated Genes in Bacteria.” Microbiology and Molecular Biology Reviews, 61(3): 319-336.
[10] Cornelis, P., Matthijs, S. and Van Oeffelen, L. (2009). “Iron uptake regulation in Pseudomonas aeruginosa.” Biometals, 22(1): 15-22. DOI: 10.1007/s10534-008-9193-0
[11] Lee, H., Kim, J., Lee, M., Park., S. and Lee, K. (2013). “Regulation of haemolysin (VvhA) production by ferric uptake regulator (Fur) in Vibrio vulnificus: repression of vvhA transcription by Fur and proteolysis of VvhA by Fur-repressive exoproteases.” Molecular Microbiology, 88(4): 813-826. DOI: 10.1111/mmi.12224
[12] Doherty, C.P. (2007). “Host-Pathogen Interactions: The Role of Iron.” Journal of Nutrition, 137(5): 1341-1344.
[13] Crowley, D.E., Wang, Y.C., Reid, C.P.P. and Szaniszlo, P.J. (1991). “Mechanisms of iron acquisition from siderophores by microorganisms and plants.” Plant and Soil, 130(1): 179-198. DOI: 10.1007/BF00011873