Trypanosoma brucei: Difference between revisions
No edit summary |
|||
| Line 1: | Line 1: | ||
{{Uncurated}} | |||
{{Biorealm Genus}} | {{Biorealm Genus}} | ||
Latest revision as of 03:38, 20 August 2010
A Microbial Biorealm page on the genus Trypanosoma brucei
Classification
Higher order taxa
Kingdom: Eukaryota
Phylum: Euglenozoa
Order: Kinetoplastida
Family: Trypanosomatidae
Genus: Trypanosoma
SubGenus: Trypanozoon
Species: Trypanosoma brucei
Species
Genus: Trypanosoma
Species: brucei
Sub-species: Trypanosoma brucei brucei, Trypanosoma brucei gambiense, Trypanosoma brucei rhodesiense, Trypanosoma brucei TREU927.
Description and Significance
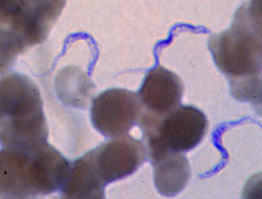
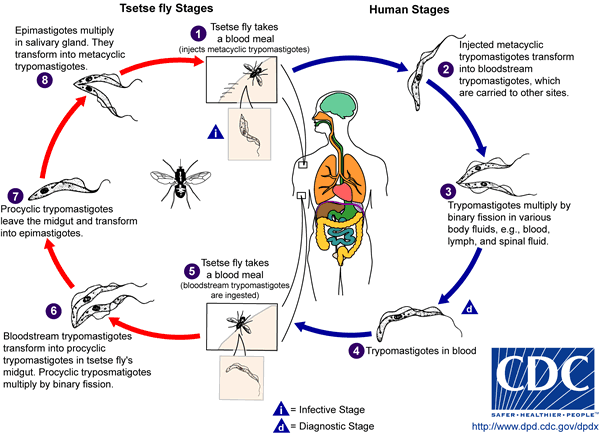
The parasitic eukaryote, Trypanosoma brucei, is a heterotrophic species from the Trypanosoma genus. It exists in two forms: an insect vector, and once inside the bloodstream, a mammalian host. T. brucei exists as its insect vector in the tsetse fly, a large, biting fly originating in Africa. Once the tsetse fly bites a mammal, the microbe enters the bloodstream where it transforms into the mammalian host form, and is then capable of mutating and invading the central nervous system, (CNS). Once inside the CNS, it has the ability to inflict African trypanosomiasis, (sleeping sickness).
The complete genome of T. brucei has been sequenced; this is important because it contains pertinent information that is used to research possible cures for African trypanosomiasis. The genome was isolated at The Institue for Genomic Research, under a project called, "The TIGR Trypanosoma brucei Genome Project," and by researchers at the Sanger Institute. Scientists used a two-part sequencing strategy. In the first phase of the project, about 37,000 sequences of 500 bp each from the ends of BAC, P1 and whole genome sheared plasmid DNA libraries were determined; the second phase of the project involved thorough and highly accurate sequencing of specific T. brucei chromosomes by iteratively selecting minimally overlapping BACs for complete sequencing.(16)
Genome structure
The genome of T. brucei has 11 megabased-size chromosomes, ranging from one to six Mbp's, which do not condense during mitosis.(6) It is predicted to have 9068 genes, with about 904 pseudogenes, and 1700 genes that are specific for T. brucei. The G+C content is 46.4%.
The genome of T. brucei also has surface antigens that allow the bacteria to escape from being noticed by the immune system.(1) T. brucei is capable of continuously changing the expression of these antigens to effectively hide from antibodies. It is thought that there are around 806 VSG's in the genome. The mechanism for antigenic variation is under parasitic genetic control. (9) The microbe can release many different pathogenic substances, that cause changes in the cytokine/prostoglandin network. These changes can activate macrophages, which then release tumor necrosis factor and nitrous oxide. These substances enhance immunosuppression, and alter the blood brain barrier so that trypansomes and inflammatory cells can invade the CNS.
The circular mitochondrial genome in T. brucei is enclosed in a single kintetoplast, which is positioned at the base of the flagellum.(7) The mitochondrial DNA in kinetoplasts is circular, and interwoven, similar to the World Olympic symbol. The circles exist in two sizes: mini and maxi. There are fewer maxi circles than there are mini circles, because of size. T. brucei has about 100 minichromosomes as well.
T. brucei also has plasmids. One of the minicircles in the kDNA was isolated, and the plasmid pTbo-1 carries the kDNA element. (17) It is supercoiled, with approximately seven to nine pTbo-1 monomer units in a head-to-tail orientation. This specific plasmid contains a sequence that allows replication and stable maintenance of extrachromosomal elements. There are only one of these specific plasmids per T. brucei cell.
The cytoskeleton of T. brucei can be divided into three main classes: microtubules, intermediate filaments, and actin microfilaments.(6)
Cell structure and metabolism
In general, the cellular structure of T. brucei is similar to all other eukaryotes. There are however, a few differences. T. brucei's cell surface has, (in addition to its surface antigens), a thick layer of proteins, called Variant Surface Glycoprotein (VSG's) genes. These allow the surface antigens to mutate, by switching variants.(2) Having over 1000 VSG genes and psuedogenes, T. brucei is able to switch variants frequently. Trascription occurs one gene at a time, from one of many telomeric VSG expression sites.(3) In order to switch an active VSG gene, DNA rearrangements must occur, to switch the old VSG gene with a new one. Using the bloodstream form of T. brucei, scientists in the Netherlands discovered that telomere exchange, thought to be rare, was indeed occuring. The scientists marked a VSG gene with a hygromycin resistance gene, allowed the gene to undergo variation, and selected switched Trypanosomes. The drug sensitivity and polymerase chain reactions (PCR), revealed that telomere exchange had taken place.(4) Interestingly enough, of the 806 VSG's in the genome of T. brucei, only about 7% of them are thought to be fully functional!
T. brucei also has unusual Citric Acid Cycles and a single large mitochondria. In the insect vector host, the Citric Acid Cycle is not used to generate energy; rather parts of the Citric Acid Cycle are suggested to be used for: acetyl-CoA transport into cytosol, degradation of proline and glutamate to succinate, and the formation of malate.(5) The Citric Acid Cycle is not functioning as a cycle itself, but parts of its pathways are being used in T. brucei.
It is very important to understand the cell's metabolism, as it is a key target for new drug synthesis. Most of the research done on T. brucei's metabolism is on the microbe's lifecycle; however, T. brucei has shown to be a the most metabolically restrictive species of the Trypanosoma genus.(6) It is thought that horizontal gene transfer from bacteria to the Tritryp lineage is the cause of this versatility. The Tritryp lineage has many essential genes that are required for the uptake/degredation of glucose. By targeting drugs to alter the pathways that use glucose, (glycolysis, the Citric Acid Cycle, or the pentose phosphate shunt), one could potentially discover new medicines for African Trypanosomiasis. Although about 50% of the genes of T. brucei have no known function, as of yet, many more biochemical pathways have yet to be discovered.
Cells usually use Type I or II synthase to undergo fatty acid synthesis. However, T. brucei has provided a third mechanism. T. brucei uses elongases to synthesize fatty acids. Other cells use elongases to make long-chain fatty acids even longer. The nature of this pathway allows synthesis of different fatty-acid end products(15), that have important roles in T. brucei's biology.
Ecology
Insect-borne diseases can have profound affects on human life, agriculture, and livestock. African trypanosomiasis affects just these. The insect carrier, is the tsetse fly; the three species of tsetse flies are mainly found in Western and Central Africa. T. brucei lives in the gut of the tsetse fly, along with two other symbionts: Sodalis glossinidius and Wigglesworthia glossinidia. (10) These other symbionts provide nutrients that the fly is either lacking in its diet, or incapable of synthesizing itself. They allow the host to live in normally unviable niches. All together, this provides a productive environment for T. brucei to thrive in.
Pathology
T. brucei resides in the tsetse flys salivary glands, as metacyclic trypomastigotes. Once the fly bites a mammal, (human or animal), the metacyclic trypomastigotes get injected into the lymphatic system, and eventually pass into the bloodstream, where they transform into bloodstream trypomastigotes.(8) There, they reproduce asexually and go through many rounds of differentiation to adapt to the drastic changes in its environment. For example, the shape of T. brucei changes from short and stumpy, to long and slender. They then travel to different regions of the body, such as: lymph, body fluid, and the CNS. Once in the CNS, T. brucei causes an irreversible demyelinating process, due to an autoimmune response.(9) It also causes swelling of the lymph nodes in the neck-the first sign of African trypanosomiasis.
It is hard to determine whether or not there are trypanosomes in the blood, because of the VSG's, (they evolve with every surface contact). A spinal tap may be necessary to test the spinal fluid, or bone marrow extractions may be needed.
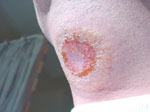
Common symptoms of African trypanosomiasis are headaches, fevers, abnormal behavior, chancres, rashes, and other symptoms similar to the effects of a seizure. The disease can induce coma, or even cause death.(8)
Treatment of T. brucei infection, or trypanosomiasis, is carried out with an array of different drugs. Prescription depends on what stage of trypanosomiasis the individual is in. For early stages, when trypansomes are only detected in the circulation, Pentamidine or Suramin are suggested. Melarsoprol may be used at all stages of trypanosomiasis, but a safer and newer drug has been developed, Eflornitine, and is used more often.
Application to Biotechnology
One of T. brucei's VSG's have an enzyme called glycosyl-phosphatidylinositol (GPI). An inositol glycan fragment from GPI was analyzed to see if it mimics the effects of insulin antilipolytic activity. This specific fragment inhibits isoproterenol-stimulated lipolysis, which insulin also inhibits. Glycan-induced antilipolysis is blocked by a low Km cAMP phosphodiesterase ihibitor, imazodan, and okadaic acid. This fragment also causes inhibition of glucose-6-phosphatase and cytosolic fructose-1,6-bisphosphatase activity (essential enzymes in glycolysis/gluconeogenesis). The direct addition of the fragment to hepatocytes caused marked inhibition of gluconeogenesis. This suggests that the direct modification of the activities of these two enzymes by the inositol glycan fragment could play a role in the inhibition of glucose output by insulin, and provide the evidence for the insulin-mimetic properties of the inositol glycan fragment. (11)
Current Research
Because T. brucei causes diseases, pathogenic parasites are constantly being studied. They are very toxic and have high resistance to current chemotherapeutics-an evergrowing issue in todays world. Today, chemotherapy is the main source of treatment against African trypanosomiasis.
A study was recently done on 18 structurally diverse bis-acridine compounds, against seven protozoan parasite species. The bioactivities and cytotoxicities of these compounds towards mammalian cells, were compared. When comparing the structure-activity, bis-acridine and acridine heterocycles had an influence on cytotoxicity. They required a specific length of approximately 10 Å. When the length was increased, the activity of the parasites decreased, as well as a decrease in cytotoxicity towards mammalian cells. Several other similar tests were carried out, such as substitutions in N alkylations with different linkers, or changing the terminal compounds. The strongest compounds recorded low-to-mid nanomolar EC50 values against T. brucei. (12) Compound bioactivity was found to not be an inhibition of trypanothione reductase, an important drug target in trypanosomatid parasites. This study demostrated the usefulness of screening bis-acridine compounds against parasitic targest, which can aid in the discovery of new chemotherapeutics-especially needed in Africa to counteract the effects of T. brucei's pathogenicity.
Other research is being conducted on heat shock therapies against T. brucei. In T. brucei, three mitogen-activated protein kinase (MAPK) pathways (which mediate developmental processes induced by stress/external signals), have been analyzed. TbMAPK4, on chromosome six, is a subclass of TbECK1. The function of TbMAPK4 is unknown, so to analyze it, null mutants of a monomorphic stock and of a pleomorphic stock were developed and tested. The various bloodstream forms of the two stocks were transformed using MAPK4-hyg. A hygromycin-resistant clone was then transformed again, and southern blot analysis showed that both copies of TbMAPK4 were deleted in one clone. This meant that null forms were able to grow normally. The optimal growth temperatures of the clones, 25 °C, was raised to 27 or 37 °C. Within 24 hours of the raise in temperature, the null mutants started to die. After 100 hours, all cells were dead. (13) The null mutants were much more heat shock sensitive than the parental lines. A possible explanation is that the null mutants might be unable to induce expression of heat shock proteins.(13) Another transformed line of TbMAPK4, HA-TbMAPK4 was tested at different temperatures. Expression of TbMAPK4 protein was always lower in cells cultured at 37 °C, but not in cells cultured at 27 °C. In fact, the cells cultured at 27 °C showed a 2 fold increase in kinase activity. Scientists believe that because the cells were still able to grow in temperatures that the insect host normally inhabited, ~25 to 27°C, this meant that TbMAPK4 was not necessary for essential growth/differentiation processes in T. brucei. (13)
New tools have been developed to amplify DNA whole genomes, from limited templates. The tools have been used to study trypanosome infections, which are characterized by low parasitemias. Multiple displacement amplification (MDA), amplifies DNA. Mouse blood samples on FTA filter cards with known numbers of T. brucei parasites were evaluated (and later Human African trypanosomiasis patients blood were tested). Data displayed a 20-fold increase in the number of PCR's possible per sample. (18) MDA allows maximal use of finite DNA samples-a valuable tool in population genetic analyses.
Continuous research on the drugs used to target trypanosomiasis is underway. T. brucei, shows resistance to tryparsamide. Many theories are being tested to explain the reason why drug resistance by T. brucei occurs. (14) Some of the theories include:
-Aquaglyeroporins
-Thiamine transporters
-High affinity pentamidine transporter (HAPT1)
-ABC transporters
References
Edited by Shannon Chan, a student of Dr. Larsen, UCSD.
(3)Taylor JE, Rudenko G. "Switching trypanosome coats: what's in the wardrobe?". 2006 Aug 14.
(8)"Parasites and health". 2005 May.