Turritopsis dohrnii: Difference between revisions
Fitzgerald3 (talk | contribs) |
Fitzgerald3 (talk | contribs) |
||
| Line 42: | Line 42: | ||
<br><br> | <br><br> | ||
===Gene Expression=== | ===Gene Expression=== | ||
Analyzing which genes are expressed during the different stages of <i>Turritopsis dohrnii’s</i> reverse development can give insights into the genetics behind their unique biological immortality. Through RNA-sequencing and transcriptome assembly, Yui Matsumoto and her team analyzed gene expression during the medusa, cyst, and polyp stages. Genes related to DNA synthesis, DNA integration, DNA transposition, DNA repair (including telomere maintenance), metabolic processes, and synthesis of nitrogen-containing cellular compounds were over-expressed in the cyst stage compared to expression in the medusa and polyp stages.<ref name=Matsumoto | Analyzing which genes are expressed during the different stages of <i>Turritopsis dohrnii’s</i> reverse development can give insights into the genetics behind their unique biological immortality. Through RNA-sequencing and transcriptome assembly, Yui Matsumoto and her team analyzed gene expression during the medusa, cyst, and polyp stages. Genes related to DNA synthesis, DNA integration, DNA transposition, DNA repair (including telomere maintenance), metabolic processes, and synthesis of nitrogen-containing cellular compounds were over-expressed in the cyst stage compared to expression in the medusa and polyp stages.<ref name=Matsumoto/> These results suggest that DNA repair and transposable elements play a key role in the reverse development and regenerative properties of <i>Turritopsis dohrnii</i>. In contrast, there was under-expression of genes related to protein synthesis, synthesis of other cellular components, cell signaling and communication, cellular division, aging and lifespan, development and cellular differentiation.<ref name=Matsumoto/> | ||
Revision as of 18:13, 9 December 2020
Introduction

Turritopsis dohrnii (previously classified as Turritopsis nutricula),[1] commonly known as the immortal jellyfish or the Benjamin Button jellyfish,[2] is a small species of jellyfish known for its unique ability to revert back to an earlier life stage through transdifferentiation.[3][4] This ontogeny reversal renders the organism virtually immortal as this process has the potential to repeat itself indefinitely, although organismal death does still occur in its natural environment.[5] Reverse development can be triggered by a variety of stressors including starvation, changes in temperature or salinity, physical damage, or age-related deterioration.[4]
Although a rare ability, reverse development has been reported in several other Cnidaria species. Some of these include Laodicea undulata,[6] Aurelia sp.,[7] Hydractinia carnea,[3] Podocoryne carnea, Eleutheria dichotoma, Cladonema sp. and Cladonema uchidai, and Perarella schneideri.[5] However, these species can only undergo reverse development during the early stages of medusa bulb development. What makes Turritopsis dohrnii unique is its ability to revert after sexual maturation.[5]
The immortal nature of Turritopsis dohrnii makes it of interest for research in aging, cancer, and regenerative medicine.
Life Cycle
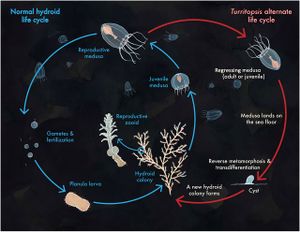
If produced sexually, the life cycle of Turritopsis dohrnii begins with planula larvae, fertilized eggs that settle on the seafloor.[8] From there, it develops into its hydroid stage. In this stage, it now takes on the form of stolons and polyps that reproduce asexually to form a colony. Immature medusae then bud off of the polyps and develop into sexually mature medusae, characterized by the presence of at least 16 tentacles,[9] within 18 to 30 days depending on the temperature of their environment.[5] The mature medusae may then release gametes that form planula larvae if they undergo fertilization, repeating the cycle. However, unlike other species, this is not the only option Turritopsis dohrnii has for restarting the cycle (Figure 2). If faced with unfavorable conditions or physical stresses (such as starvation, changes in temperature or salinity, physical damage, or age-related deterioration)[4] in the immature medusa stage, the organism will skip the maturation and sexual reproductive stages and revert back to the hydroid stage. All mature medusae will spontaneously regress back to the hydroid stage as well, whether or not they face adverse environmental conditions.[5] This reverse development is likely the result of senescence-related stress.
Reverse Development
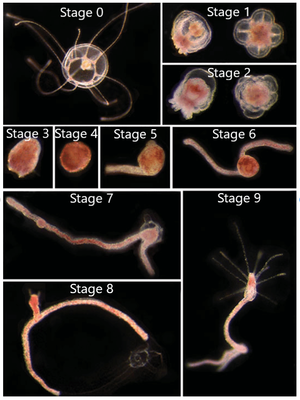
The reverse development process can begin from either the immature or mature medusa stage (Figure 3, Stage 0). The first steps in the process is the reduction of medusa features to create a “ball-like” stage (Figure 3, Stages 1-4), also commonly known as a cyst stage.[3] However, if the organism is in the transitional stage between maturity, characterized by the presence of 13-15 tentacles, there is a 20%-40% chance that the medusa will develop directly into the hydroid stage, skipping the cyst stages.[5] The next step in reverse development is stolon formation in which branches form off the ball-like tissue (Figure 3, Stages 5-7). This step is followed by the formation of a polyp bud (Figure 3,, Stage 8) and finally adult polyp formation (Figure 3, Stage 9).[3] From here, the polyp may asexually reproduce and bud off new medusae. In this sense, Turritopsis dohrnii is “immortal” as it doesn’t die at the end of its lifecycle; it simply starts anew from an earlier developmental life stage.
Genetics
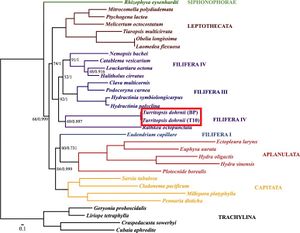
Mitochondrial DNA
A complete analysis of the mitochondrial DNA in Turritopsis dohrnii was done by A.A. Lisenkova and her team at the Russian Academy of Sciences in 2017. The mitochondrial DNA of Turritopsis dohrnii was made up of 15,425 bp that comprised 13 protein-coding genes, two tRNAs, and two rRNA subunits.[10] None of these genes seemed to reveal the secrets to the organism’s death defying abilities. The protein-coding genes found were all associated with oxidative phosphorylation and respiration,[10] typical of mitochondrial DNA. Because of the limited nature of mitochondrial DNA, not many insights could be made from the analysis, however, the results did solidify Turritopsis dohrnii’s phylogenetic placement. rrnS and protein-coding sequence analysis placed Turritopsis dohrnii and R. octopunctata as distinct group from their Filifera III + Filifera IV clade which was consistent with previous analyses (Figure 4).[10]
Gene Expression
Analyzing which genes are expressed during the different stages of Turritopsis dohrnii’s reverse development can give insights into the genetics behind their unique biological immortality. Through RNA-sequencing and transcriptome assembly, Yui Matsumoto and her team analyzed gene expression during the medusa, cyst, and polyp stages. Genes related to DNA synthesis, DNA integration, DNA transposition, DNA repair (including telomere maintenance), metabolic processes, and synthesis of nitrogen-containing cellular compounds were over-expressed in the cyst stage compared to expression in the medusa and polyp stages.[8] These results suggest that DNA repair and transposable elements play a key role in the reverse development and regenerative properties of Turritopsis dohrnii. In contrast, there was under-expression of genes related to protein synthesis, synthesis of other cellular components, cell signaling and communication, cellular division, aging and lifespan, development and cellular differentiation.[8]
Microbial Relationships
Research on the microbiota of Turritopsis dohrnii has not been done, however, there is one interesting microbial relationship that is essential to Hydrozoan development. In order to progress from the planula larvae stage to the hydroid stage, Turritopsis dohrnii require an external signal from marine bacteria.[3] The exact species of bacteria that induces metamorphosis in Turritopsis dohrnii has not been studied, however, there is research on the bacterial inducers for Hydractinia echinta, a closely related Hydrozoan. In Hydractinia echinta, Alteromonas espejiana[11] and other Alteromonas species[12] are known to be strong inducers, but other common marine bacteria, including E. coli can also induce metamorphosis.[12] The presence of these bacterial signals are essential to Hydrozoan development as the planula larvae will not undergo metamorphosis without them.[11]
Medical Potential
Conclusion
Overall text length should be at least 1,000 words (before counting references), with at least 2 images. Include at least 5 references under Reference section.
References
- ↑ Schuchert P. Revision of the European athecate hydroids and their medusae (Hydrozoa, Cnidaria): families Oceanidae and Pachycordylidae. Revue suisse de Zoologie. 2004 Jun 1;111(2):315-70. Available from: https://www.semanticscholar.org/paper/Revision-of-the-European-athecate-hydroids-and-and-Schuchert/e3d3dce80157e5ee98ecbe0bbe7d35eb36cbe82b?p2df
- ↑ Than K. "Immortal" Jellyfish Swarm World's Oceans [Internet]. National Geographic. 2009. Available from: https://www.nationalgeographic.com/animals/2009/01/immortal-jellyfish-swarm-oceans-animals/
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 Schmich J, Kraus Y, De Vito D, Graziussi D, Boero F, Piraino S. Induction of reverse development in two marine Hydrozoans. Int J Dev Biol. 2007 Feb 1;51:45–56. Available from: https://www.researchgate.net/publication/6617831_Induction_of_reverse_development_in_two_marine_Hydrozoans
- ↑ 4.0 4.1 4.2 Piraino S, De Vito D, Schmich J, Bouillon J, Boero F. Reverse development in Cnidaria. Canadian Journal of Zoology. 2004 Nov 1;82(11):1748-54. Available from: https://www.researchgate.net/profile/Stefano_Piraino/publication/249542511_Reverse_development_in_Cnidaria/links/004635220749d81dc3000000/Reverse-development-in-Cnidaria.pdf
- ↑ 5.0 5.1 5.2 5.3 5.4 5.5 Piraino S, Boero F, Aeschbach B, Schmid V. Reversing the life cycle: medusae transforming into polyps and cell transdifferentiation in Turritopsis nutricula (Cnidaria, Hydrozoa). The Biological Bulletin. 1996 Jun 1;190(3):302-12. Available from: https://www.journals.uchicago.edu/doi/pdfplus/10.2307/1543022?casa_token=VCv_136r__UAAAAA%3ARVFGUXoAfbOsfuX2x2l8RiTdYNa4mgvDPrnVFQEo5KUbmRat544_LD8iRbOzKJjDNuSi38-gjDo7&
- ↑ De Vito D, Piraino S, Schmich J, Bouillon J, Boero F. Evidence of reverse development in Leptomedusae (Cnidaria, Hydrozoa): the case of Laodicea undulata (Forbes and Goodsir 1851). Marine Biology. 2006 May 1;149(2):339-46. Available from: https://link.springer.com/article/10.1007%2Fs00227-005-0182-3
- ↑ He J, Zheng L, Zhang W, Lin Y. Life cycle reversal in Aurelia sp. 1 (Cnidaria, Scyphozoa). PloS one. 2015 Dec 21;10(12):e0145314. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4687044/
- ↑ 8.0 8.1 8.2 Matsumoto Y, Piraino S, Miglietta MP. Transcriptome characterization of reverse development in Turritopsis dohrnii (Hydrozoa, Cnidaria). G3: Genes, Genomes, Genetics. 2019 Dec 1;9(12):4127-38. Available from: https://www.g3journal.org/content/ggg/9/12/4127.full.pdf
- ↑ Martell L, Piraino S, Gravili C, Boero F. Life cycle, morphology and medusa ontogenesis of Turritopsis dohrnii (Cnidaria: Hydrozoa). Italian Journal of Zoology. 2016 Jul 2;83(3):390-9. Available from: https://www.tandfonline.com/doi/full/10.1080/11250003.2016.1203034
- ↑ 10.0 10.1 10.2 Lisenkova AA, Grigorenko AP, Tyazhelova TV, Andreeva TV, Gusev FE, Manakhov AD, Goltsov AY, Piraino S, Miglietta MP, Rogaev EI. Complete mitochondrial genome and evolutionary analysis of Turritopsis dohrnii, the “immortal” jellyfish with a reversible life-cycle. Molecular phylogenetics and evolution. 2017 Feb 1;107:232-8. Available from: https://www.sciencedirect.com/science/article/pii/S1055790316303359?casa_token=zzQIehLx3UwAAAAA:zRnlJmhcU93bxbb4my_kh5LL3z4CLEGjW3WA-aL-UZSl1sa-sQF68dnUdZefETMpK13fAWS5bP8
- ↑ 11.0 11.1 Leitz T, Wagner T. The marine bacterium Alteromonas espejiana induces metamorphosis of the hydroid Hydractinia echinata. Marine Biology. 1993 Feb 1;115(2):173-8. Available from: https://link.springer.com/article/10.1007/BF00346332
- ↑ 12.0 12.1 Kroiher M, Berking S. On natural metamorphosis inducers of the cnidarians Hydractinia echinata (Hydrozoa) and Aurelia aurita (Scyphozoa). Helgoland Marine Research. 1999 Nov;53(2):118-21. Available from: https://hmr.biomedcentral.com/track/pdf/10.1007/s101520050014.pdf?site=http://hmr.biomedcentral.com
Edited by Bailey Fitzgerald, student of Joan Slonczewski for BIOL 116 Information in Living Systems, 2020, Kenyon College.
