User:S4338571: Difference between revisions
No edit summary |
|||
| Line 13: | Line 13: | ||
==Description and significance== | ==Description and significance== | ||
[[File:fmicb-07-00053-g002.jpg|thumb|alt=Puzzle globe|Morphology of ''P. gingivalis'' on horse blood agar. Accumulation of hemin causes the production of black-pigmented colonies<sup>[[#References|[3]]]</sup>]] | [[File:fmicb-07-00053-g002.jpg|thumb|alt=Puzzle globe|Morphology of ''P. gingivalis'' on horse blood agar. Accumulation of hemin causes the production of black-pigmented colonies.<sup>[[#References|[3]]]</sup>]] | ||
''Porphyromonas gingivalis'' is a gram-negative, obligate anaerobic, non-motile, non-spore-forming microorganism and is one of the predominant human oral microbiota. <sup>[[#References|[1]]]</sup> This rod-shaped, black-pigmented, asaccharolytic and highly proteolytic bacterium cannot grow in the existence of bile (20%) and on rabbit blood agar plates, they have an average size of diameter below 1.5 μm, living individually from each other. <sup>[[#References|[1]]]</sup>,<sup>[[#References|[2]]]</sup> It is often found in a deep periodontal pocket, human subgingival plaque, living along with approximately other >500 species of bacteria. <sup>[[#References|[3]]]</sup> | ''Porphyromonas gingivalis'' is a gram-negative, obligate anaerobic, non-motile, non-spore-forming microorganism and is one of the predominant human oral microbiota. <sup>[[#References|[1]]]</sup> This rod-shaped, black-pigmented, asaccharolytic and highly proteolytic bacterium cannot grow in the existence of bile (20%) and on rabbit blood agar plates, they have an average size of diameter below 1.5 μm, living individually from each other. <sup>[[#References|[1]]]</sup>,<sup>[[#References|[2]]]</sup> It is often found in a deep periodontal pocket, human subgingival plaque, living along with approximately other >500 species of bacteria. <sup>[[#References|[3]]]</sup> | ||
| Line 29: | Line 29: | ||
==Cell structure and metabolism== | ==Cell structure and metabolism== | ||
[[File:fmicb-07-00053-g004.jpg|thumb|alt=Puzzle globe|Overview of the ''P. gingivalis'' lipopolysaccharide structure (LPS) of outer membrane<sup>[[#References|[3]]]</sup>]] | [[File:fmicb-07-00053-g004.jpg|thumb|alt=Puzzle globe|Overview of the ''P. gingivalis'' lipopolysaccharide structure (LPS) of outer membrane.<sup>[[#References|[3]]]</sup>]] | ||
The cell structure of ''Porphyromonas gingivalis'' consists of capsules, fimbriae, lipopolysaccharide (LPS) and outer membrane proteins. <sup>[[#References|[3]]]</sup> Capsules are important for the initial adherence to the host’s teeth, also is vital to provide resistance to the flow of saliva. <sup>[[#References|[3]]]</sup> In vivo experiment has shown that encapsulated strain of ''P. gingivalis'' is more virulent compared to non-encapsulated strain which only caused local infection when infected in mice. This is because the non-encapsulated strains are more susceptible to phagocytosis or can be easily killed by dendritic cells and macrophages while encapsulated strains are able to modulate the host’s immune response, lowering the production of cytokines interleukin-1 (IL-1), IL-6, and IL-8 by fibroblasts. <sup>[[#References|[14]]]</sup> | The cell structure of ''Porphyromonas gingivalis'' consists of capsules, fimbriae, lipopolysaccharide (LPS) and outer membrane proteins. <sup>[[#References|[3]]]</sup> Capsules are important for the initial adherence to the host’s teeth, also is vital to provide resistance to the flow of saliva. <sup>[[#References|[3]]]</sup> In vivo experiment has shown that encapsulated strain of ''P. gingivalis'' is more virulent compared to non-encapsulated strain which only caused local infection when infected in mice. This is because the non-encapsulated strains are more susceptible to phagocytosis or can be easily killed by dendritic cells and macrophages while encapsulated strains are able to modulate the host’s immune response, lowering the production of cytokines interleukin-1 (IL-1), IL-6, and IL-8 by fibroblasts. <sup>[[#References|[14]]]</sup> | ||
Latest revision as of 17:59, 22 September 2016
Muhammad Irfan Zulkifle Bench E 23/9/2016 [1]
Classification
Higher order taxa
Bacteria (Kingdom) – Fibrobactere-Chlorobi-Bacteroidetes, FCB Group (Domain) – Bacteroidetes (Phylum) – Bacteroidia (Class) – Bacteroidales (Order) – Porphyromonadaceae (Family) – Porphyromonas (Genus)
Species
Porphyromonas gingivalis, Strain W83
Description and significance
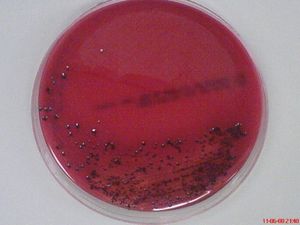
Porphyromonas gingivalis is a gram-negative, obligate anaerobic, non-motile, non-spore-forming microorganism and is one of the predominant human oral microbiota. [1] This rod-shaped, black-pigmented, asaccharolytic and highly proteolytic bacterium cannot grow in the existence of bile (20%) and on rabbit blood agar plates, they have an average size of diameter below 1.5 μm, living individually from each other. [1],[2] It is often found in a deep periodontal pocket, human subgingival plaque, living along with approximately other >500 species of bacteria. [3]
P. gingivalis is a primary causative pathogen that contributed to a chronic periodontitis, a disease that is characterized by the demolition of tooth-supporting tissues, affecting about 50% of population >30 years of age in the United States and globally, it affects about 10-15% adult populations. [1],[4] The disease usually started as an acute gingival tissue inflammation, but then may advance to a creation of teeth pocket which may cause loss of teeth if it is not treated. [3] Periodontitis is more susceptible among patient acquiring systemic diseases. [5],[6]
Periodontitis has been linked with cardiovascular diseases such as coronary artery disease, heart attack and stroke. [3],[7] Other than periodontitis, P. gingivalis has also been associated with pulpal infection, oral abscesses and it was also detected in women with bacterial vaginosis which may cause burning with urination. [8],[9],[10] The bacteria of virulent strain, W83 was first discovered in the 1950s at Bonn, Germany by H.Werner, obtained from an undocumented oral disease and then in 1960s, it was brought by Madeleine Sebald to The Pasteur Institute. [11] P. gingivalis has been cultured and was available at American Type Culture Collection. [12]
Although there have been numerous studies done to explain the mechanism of virulence factors secreted by P. gingivalis and how they interact with the host, investigating a gene or protein in isolation without considering other molecular networks is not truly insightful because in the actual in vivo environment, the genes may work as a system, hence may interact differently than a single virulence factor to the host cells. Adding to that, rather than working alone, P. gingivalis is also likely to interact with other microbes to survive in the harsh environment of the periodontal pocket. [3] Therefore, further research needs to be performed to improve our understanding of the interaction between periodontal bacteria and host cells at the molecular and cellular level so that effective approaches can be implemented to control the disease caused by this bacterium.
Genome structure
The genome size of circular strain W83 (GenBank: AE015924.1) is 2,343,479 bp containing 4 ribosomal operons (5S-23S-tRNAAla-tRNAIle-16S), 2 structural RNA genes and 53 specific amino-acid tRNA genes, with an average G+C content of 48.3% and total ORF of 1990 (covered the complete genome by 85%). [11],[13] 54% of the ORF consisted of biological role categories, 10.5% have an unknown function, 9.2% were conserved hypothetical proteins/domain proteins while the other 26.3% were hypothetical proteins.[11] Repetitive elements such as DNA repeats (clustered regularly interspaced short palindromic repeats and uninterrupted direct repeats) and transposable components (insertion sequence elements and miniature inverted-repeat transposable elements) made up 6% of the whole genome.[11] The strain did not contain other classes of dispersed repetitive DNA sequence elements which include ERIC and REP elements.[11]
Cell structure and metabolism
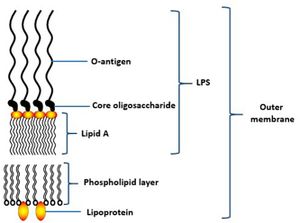
The cell structure of Porphyromonas gingivalis consists of capsules, fimbriae, lipopolysaccharide (LPS) and outer membrane proteins. [3] Capsules are important for the initial adherence to the host’s teeth, also is vital to provide resistance to the flow of saliva. [3] In vivo experiment has shown that encapsulated strain of P. gingivalis is more virulent compared to non-encapsulated strain which only caused local infection when infected in mice. This is because the non-encapsulated strains are more susceptible to phagocytosis or can be easily killed by dendritic cells and macrophages while encapsulated strains are able to modulate the host’s immune response, lowering the production of cytokines interleukin-1 (IL-1), IL-6, and IL-8 by fibroblasts. [14]
Fimbriae are thin, about 3-25μm long structures that extend out of bacterial outer membrane.[3] P. gingivalis expresses two types of fimbriae; fimbrillin (FimA), major and long fimbriae encoded by and Mfa protein, minor and short fimbriae.[3] Fimbriae are important for binding and invasion of host cells, also vital for adherence to various oral molecules such as commensal bacteria (for example streptococci).[15] Other than long and short fimbriae, there are also accessory fimbriae; Fim C, D and E which have a role in matrix proteins binding and interaction with CXC-chemokine receptor 4.
Lipopolysaccharides (LPS) is a vital structure of the bacterial outer membrane and usually very large in size, >10kDa.[3] It consists of a distal polysaccharide (O-antigen), a core oligosaccharide and a hydrophobic domain (lipid A or endotoxin) (3). The most inner component, endotoxin has heterogeneous acylation pattern which is dynamic depending on environmental surrounding and is able to affect immune signalling of the host, thus increasing the likelihood of survival.[3] LPS plays a vital role in maintaining the cellular and structural integrity of the bacteria, regulating the entry of hydrophobic molecules and toxic chemicals, also involves in folding and insertion of outer membrane protein.[3] Furthermore, LPS is a key component that leads to periodontitis as it is able to activate the host inflammatory responses, such as the production of IL-1, eventually causing the destruction of periodontal tissue.[16]
The cell envelope of P. gingivalis consists of inner and outer membrane, separated by periplasm containing peptidoglycan layer.[3] The cell membrane acts as a selective barrier that only allows certain substances to enter the cell, hence providing protection to the cell.[3] OM proteins which include lipoproteins and integral proteins involved in most specific recognition processes of the bacteria and also important for the formation of periodontal biofilms.[3] Porins and OmpA-like proteins are the most abundant OM proteins. LptO and PG534 are another examples of OM proteins. LptO is important for O-deacylation of LPS, vital for cell attachment while PG534 is important to increase the activities of gingipains.[3] Gingipains are key components of the cell for taking up nutrients, for example, degrades the host’s haemoglobin to acquire iron and haem, also degrades the host’s albumin serum for obtaining enough carbon and nitrogen source.[17] Other than that, gingipains involve in degrading the signals of immune response.[18]
P. gingivalis is found to create a synergistic biofilm with Treponema denticola, forming a symbiotic relationship, particularly for utilization of nutrients and also pathogenicity in periodontal disease.[19] P. gingivalis is usually found beneath the spirochete layer while T. denticola is mostly observed in the surface layers of subgingival plaque.[19] Compared in monospecies biofilms, the growth of P. gingivalis is more enhanced in synergistic biofilms as T. denticola produced an important compound, succinate which facilitates its survival.[19]
Ecology
P. gingivalis is an obligate anaerobe and predominantly found in subgingival sulcus, the space between the free gingiva and the tooth surface.[3],[20] It is also found in a deep periodontal pocket where the availability of sugar is usually very low, hence for energy production, P.gingivalis is able to ferment amino acids.[3]
P. gingivalis is usually a secondary colonizer of a dental plaque and the colonization is mediated by saliva which serves as a platform for its transmission and initial entrance into the oral milieu.[3],[20] The tooth surfaces which are usually coated with salivary pellicle provides attachment for the bacterial fimbriae to resist salivary flow.[20] Eventually, P. gingivalis can reach the subgingival crevice by proliferation with the help of primary colonizer such as Streptococcus gordonii that serves as a site of attachment.[20] Streptococcus gordonii is a facultative anaerobe which can decrease the oxygen levels permissive for the survival of obligate anaerobic.[20] Other than that, the bacteria are also found in the upper gastrointestinal tract, women with bacterial vaginosis, respiratory tract and colon.[10]
To stay alive in the crevice, the microbe needs to interact with the host’s epithelial cells and neutrophils.[20] P. gingivalis adheres, actively invades and replicates within epithelial cells to prevent recognition and surveillance of the immune system.[20] To survive and persist longer in the host’s epithelial cells, P. gingivalis has the genes that encode for nucleoside diphosphate kinase, an enzyme that suppresses ATP-induced apoptosis, hence preventing epithelial cell apoptosis.[20] Also, by invading the cell, it is able to suppress the production of IL-8 by epithelial cells, leading to suppression or delay of neutrophil influx.[20] P. gingivalis is able to resist to environmental oxidative stress generated by neutrophil and oxidative killing by phagocytes by the role played by proteins rubrerythrin and alkyl hydroperoxide reductase.[21]
Pathology
The major etiologic agent that leads to chronic periodontitis is Porphyromonas gingivalis, found in 85.75% subgingival plaque samples from patients acquiring chronic periodontitis.[3] Generally, periodontal disease refers to the oral inflammatory infections caused by oral pathogens, causing destruction of tooth supporting tissues.[3] Periodontal disease can be categorized into two groups; gingival diseases and periodontitis, and the severity of the disease can be ranged from mild, reversible inflammation to long-term demolition of connective tissues, leading to the loss of teeth.[3]
Gingival disease usually referred as inflammation of gingival tissues, manifested only by mild tissues’ redness and swelling, also not affecting the attachment of teeth.[3] Meanwhile, periodontitis involves the infection of periodontal ligament and alveolar bone due to the irreversible plaque-induced inflammation of the periodontal tissues.[3] The usual clinical manifestation of the disease is the formation of periodontal pocket and subgingival plaque.[3] The disease is most likely to occur to the individuals acquiring systemic diseases.[3]
Periodontal disease has been associated with cardiovascular diseases.[22] The mechanism by which the bacteria caused the disease was unclear, but it was postulated that the bacteria is able to enter the bloodstream, causing inflammation which increases the amount of plaque that will dilate the arteries.[7] The disease has also been associated with the occurrence of pre-term low birth weight babies, mainly due to the infection in the placental unit.[23] Apart of that, a recent study indicates a possibility that P. gingivalis is able to cause rheumatoid arthritis (RA), which is characterized by disease-specific autoimmunity to a post-translational modification of arginine residues (citrullinated proteins), mediated by of peptidylarginine deiminases of P. gingivalis.[16]
Application to biotechnology
The advancement in bioinformatics has made it possible to identify new targets for vaccine. A recent study has identified 120 genes that served as potential candidates.[24] The genes cloned for expression in Escherichia coli and screened with P. gingivalis antisera before being tested in mice.[24] Two recombinant proteins, PG32 and PG33 (outer membrane porin, OprF) has demonstrated significant protection which is about 70%.[24] For further studies, they suggested the combination of PG32 and PG33 in a single vaccine which may increase the protection level.
Another study has utilized the use of arginine-specific cysteine proteinase, RgpA. They found that the vaccine triggers the production of high-level serum antibodies which is able to diminish the proteolytic activity of RgpA and RgpB, also prevent the binding of P. gingivalis to a type I collagen sponge.[25] Other than OMPs and Gingipains (RgpA), the suggested production of P. gingivalis vaccine involved the use other surface antigens such as capsules.[26] Adding to that, the focus of the administration mode by oral/nasal vaccination should be on triggering the mucosal immune response as the induction of IgA is preferable compared to the production of IgG by B cells may lead to prolong alveolar bone loss.[26]
Current research
One of the current studies suggested a possible mechanistic link between periodontal disease and tumor development in the oral cavity, by the action of P. gingivalis to disrupt the control system of kallikrein-like proteinase (KLKs).[27] The virulence factor secreted by the bacteria, gingipains is thought to destabilize the regulation of KLK by inactivating the epithelial specific inhibitor of kallikreins, called as serine protease inhibitor of kazal-type 6 (SPINK6).[27] Another research conducted has found the presence of P. gingivalis in the esophagus for the first time in the patients with esophageal cancer, particularly esophageal squamous cell carcinoma (ESCC).[28] The result suggested the possible association between the infection by P. gingivalis and the progression of ESCC, though the mechanism is unknown. The study also provided an indication that the presence of P. gingivalis can be utilized as the disease’s biomarker, also suggested that the removal of a common oral pathogen could possibly reduce the overall burden of ESCC.[28]
Apart from that, current research has demonstrated that the use of prenyl flavonoids, a natural product from the medical plant is able to inhibit the growth of P. gingivalis and the activity of gingipains, also preventing the formation of biofilm, hence suggesting its potential for the use of periodontitis treatment.[29]
References
1. Porphyromonas gingivicanis strain ATCC 55562
5. Oxidative stress and periodontal disease in Down Syndrome
12. Porphyromonas gingivalis (Coykendall et al.) Shah and Collins
- ↑ MICR3004
This page is written by Muhammad Irfan Zulkifle for the MICR3004 course, Semester 2, 2016
