User:S4355945
Rothia dentocariosa
Alexandria Chin, Bench C, 23 September 2016
Classification
Higher order taxa
Bacteria – Terrabacteria group – Actinobacteria – Actinobacteria – Micrococcales – Micrococcaceae – Rothia [1]
Species
Rothia dentocariosa
Type strain: strain ATCC 17931= CCUG 35437 = CIP 81.83 = DSM 43762 = DSM 46363 = IFO (now NBRC) 12531 = JCM 3067 = NCTC 10917 = NRRL B-8017. [2]
Description and Significance
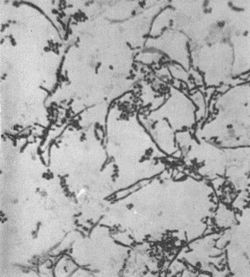
Rothia dentocariosa was first discovered by Onisi in 1949, from dental plaques and caries in humans. [4] From his study of the organism he proposed it be named A. dentocariosus because of the morphological similarities with Actinomyces species found in the mouth. His study of the isolate showed that it was a highly pleomorphic, facultative anaerobic organism with both coccoid and branched filament structures. [3] In 1957 while studying carious dentine, Roth found a similar group of organisms and believed it to be the same as Onisi’s isolate. However, she found the organism to grow under aerobic conditions and therefore suggested the name N. dentocariosus. [3][5]
Davis and Freer (1960) were unaware of Roth and Onisi’s studies and described a similar group of isolates from the human mouth. They initially suggested the name N. salivae but further studies of the cell wall showed that this organism did not belong in the Nocardia genus as it did not contain diaminopimelic acid, which is a characteristic of Nocardia genus. Georg and Brown in 1967 created the genus Rothia in the Actinomycetaceae family to accommodate this organism. Rothia was named after Genevieve D. Roth, as a tribute to her work with the organism. [3] In 1997, Stackebrandt et al. moved Rothia to the Micrococcaceae family where it shares signature nucleotides in the 16S rDNA sequence with other genera in the family. [6]
R. dentocariosa is a normal inhabitant of the oral cavity, a study showed over 30% of healthy individuals have this organism in their mouth. [4] It is present in saliva and is frequently isolated from supragingival plaque and respiratory tract. [7] It is not commonly isolated from clinical specimens but it has been isolated from blood, spinal fluids and abscesses. [3]
R. dentocariosa is a gram-positive, non-motile, non-spore forming, pleomorphic organism, that can present in coccoid and/or filamentous forms. [8][9] It has a highly variable microscopic morphology. [3] Morphology differs between young and mature colonies. Young colonies are 1mm in diameter, have entire edges and are smooth to a slight rough with a mucoid consistency. While, mature colonies can be 1-4mm, raised with a rough and cerebriform surface irregular or scalloped edges. [3] Laboratory growth of R. dentocariosa requires an enriched medium, such as TSA, [3] and organic nitrogen, however carbon dioxide is not necessary. [10]
Studying this organism and determining specific identifiers is important as Rothia is often overlooked, as a pathogen, [7] due to its similarities in morphology to Actinomyces and Nocardia, which have pathogenic members. [3] Isolates of R. dentocariosa have been found in periodontal lesions and associated with several cases of infective endocarditis. [9]
Genome structure
R. dentocariosa strain ATCC 17931 has a circular genome with 2, 506, 025 bp and is made up of 1 chromosome. It has a total of 2, 126 genes with 2, 040 coding DNA sequences and encodes 2, 213 proteins. [11] From a genome annotation, 62 RNA sequences were identified, 10 rRNAs (4, 3 and 3 of 5S, 16S and 23S respectively), 49 tRNAs and 3 ncRNAs. [12]
Cell structure and Metabolism
As R. dentocariosa is a gram-positive organism, its cell wall has a defined layer of peptidoglycan of the A3-alpha type, which has D-alanine in position 4, L-lysine in position 3 and an inter-peptide bridge of L-alanine. [10] Analysis of the cell wall showed that the sugars galactose, fructose, glucose and ribose are involved. [8] R. dentocariosa is a non-motile organism and does not have aerial mycelium, making it non-sporogenic. [9]
Coaggregation bridging is critically important for the development of microbial plaque and it was observed that Bacteroides intermedius coaggregates with R. dentocariosa in the oral microflora. Bacteroides species is observed several days after on freshly cleaned tooth surface, which recognize microcolonies made by Streptococcus sanguis. R. dentocariosa attaches to the B. intermedius and is prevented from washing out of the oral cavity by it’s adherence to the tooth surface, via S. sanguis. [13]
Laboratory testing of the organism’s metabolism showed activity of catalase production, nitrate and nitrite reduction, esculin hydrolysis and acid production from glucose, sucrose, maltose, salicin and glycerol. This can be compared to Nocardia species, which can oxidise sugars but does not ferment them into acid. [3] R. dentocariosa’s glucose fermentation also makes it unsuitable for the Actinomyces species as it produces lactic and acetic acid when Actinomyces produce succinate. [7]
Ecology
R. dentocariosa is a facultative anerobic organism found in the human oropharynx and upper respiratory tract. [14][15] Its respiratory needs make it distinctive from Actinomyces, as Actinomyces thrive in anaerobic environments while R. dentocariosa grows better in an aerobic environment, in addition it differs in morphology when cultured in aerobic conditions. [3] When cultured under anaerobic conditions, development of the filamentous form is stimulated; this form shows a loss of gram positivity. [3]
A commensalism relationship between the host and bacteria is observed. R. dentocariosa benefits from the human oral cavity, in nutrients and an optimal environment for growth, while the host goes unharmed unless immunocompromised.
Pathology
R. dentocariosa has been found particularly in patients with periodontal disease, showing that it is an opportunistic pathogen. [7] Infective endocarditis is rare but the most predominant and severe clinical manifestation of R. dentocariosa, and case studies also suggest that oral hygiene is a factor. Cases showed that a pre-existing abnormality of the aortic or mitral valve is required for infection. [9] There have been affiliations in cases of pneumonia and bacteremia, which can lead to systemic disease. [9][16] Binder et al. [9] suggested a combination of rifampin and a beta-lactam antibiotic for treatment of R. dentocariosa endocarditis. Additionally, Salamon and Prag [7] found that the most efficient beta-lactam is ceftriaxone, which should be used concurrently with rifampin if there is complicated infection, this is considered the first choice treatment. [9] However, Ferraz et al. [4] recommended penicillin and if needed aminoglycoside. It appeared that different cases presented in each article had varied susceptibility to antibiotics; each suggested treatment was successful for the particular patient. An antibiotic susceptibility test, tablet diffusion test, is conducted to determine which antibiotic is best used for the isolated strain from a clinical sample. [7] Aminoglycosides have a considerable amount of variability in their efficacy depending on the isolate of R. dentocariosa. [9]
Application to Biotechnology
R. dentocariosa encodes 2 non-redundant protein sequences that can be used as drug targets, cell division protein, FtsZ, which is a metalloendopeptidase and ribonucleotide-diphosphate reductase subunit beta. [11] Cell division protein, FtsZ, [17] is 403 amino acids in length; its role is to assemble at the site of cell division forming a ring. FtsZ is the prokaryotic homolog of tubulin in eukaryotes. This is a potential drug target as cell division is essential for R. dentocariosa to grow to the sufficient amount resulting in infections. Ribonucleotide-diphosphate reductase (RNR) [18] is an enzyme, encoded by 325 amino acids, that catalyses the reaction of ribonucleotides to deoxyribonucleotides, which are involved in the synthesis of DNA. RNR has a critical role in maintaining DNA to cell mass during cell division and DNA repair; this ratio is maintained by regulating the rate of DNA synthesis. Again, this can be used as a drug target because if the regulation of DNA synthesis is altered or turned off the organism will not be able to survive.
Current Research
Recent articles have been reviews of specific cases presented in hospital settings, which look at multiple patients presented over a short time period, such as a year. These reports identify unsuccessful diagnoses and investigate what was overlooked in the differential and how successful a diagnosis could be made for R. dentocariosa. [7][9][14]
Clinical testing of patient samples can take days to grow cultures and therefore limiting the efficiency and effectiveness of treatment. Specific selective media has been researched, RDSM, which is highly selective for R. dentocariosa and can be used for isolation in clinical samples, as it has a recovery of 93.8%. This new medium can assist in the determination of R. dentocariosa distribution and role in the human oral cavity. [15]
Biofilm growth is not well understood for this organism, though there have been recent studies showing that dental plaque biofilm development may require cross feeding and cooperation between organisms. [19][20]
References
- ↑ NCBI Taxonomy Browser
- ↑ List of Prokaryotic Names with Standing in Nomenclature
- ↑ 3.00 3.01 3.02 3.03 3.04 3.05 3.06 3.07 3.08 3.09 3.10 3.11 Brown, J.M., Georg, L.K., and Waters, L.C. (1969) Laboratory Identification of Rothia dentocariosa and Its Occurrence in Human Clinical Materials. Applied Microbiology 17(1), 150-156.
- ↑ 4.0 4.1 4.2 Ferraz, V., McCarthy, K., Smith, D., Koornhof, HJ. (1998) Rothia dentocariosa endocarditis and aortic root abscess. Journal of Infection 37(3), 292-295.
- ↑ Roth, G. D., & Thurn, A. N. (1962) Continued study of oral Nocardia. J Dent Res 41, 1279-1292.
- ↑ Stackebrandt, E., Rainey, F. A., & Ward-Rainey, N. L. (1997) Proposal for a new hierarchic classification system, Actinobacteria classis nov. International Journal of Systematic and Evolutionary Microbiology 47(2), 479-491.
- ↑ 7.0 7.1 7.2 7.3 7.4 7.5 7.6 Salamon, S. A., & Prag, J. (2002) Three cases of Rothia dentocariosa bacteraemia: frequency in Denmark and a review. Scandinavian journal of infectious diseases 34(2), 153-157.
- ↑ 8.0 8.1 Von Graevenitz, A. (2004) Rothia dentocariosa: taxonomy and differential diagnosis. Clinical microbiology and infection 10(5), 399-402.
- ↑ 9.0 9.1 9.2 9.3 9.4 9.5 9.6 9.7 9.8 Binder, D., Widmer, U., Opravil, M., Krause, M., & Zbinden, R. (1997) Native and prosthetic valve endocarditis caused by Rothia dentocariosa: Diagnostic and therapeutic considerations. Infection 25(1), 22-26.
- ↑ 10.0 10.1 Georg, L. K., & Brown, J. M. (1967) Rothia, gen. nov. an aerobic genus of the family Actinomycetaceae. International Journal of Systematic and Evolutionary Microbiology 17(1), 79-88.
- ↑ 11.0 11.1 Pathosystems Resource Integration Center
- ↑ Rothia dentocariosa ATCC 17931, complete genome
- ↑ Kolenbrander, P. E., Andersen, R. N., & Holdeman, L. V. (1985) Coaggregation of oral Bacteroides species with other bacteria: central role in coaggregation bridges and competitions. Infection and immunity 48(3), 741-746.
- ↑ 14.0 14.1 Ramanan, P., Barreto, J. N., Osmon, D. R., & Tosh, P. K. (2014) Rothia bacteremia: a 10-year experience at Mayo Clinic, Rochester, Minnesota. Journal of clinical microbiology 52(9), 3184-3189.
- ↑ 15.0 15.1 Uchibori, S., Tsudukibashi, O., Goto, H., Kobayashi, T., & Aida, M. (2012) A novel selective medium for the isolation and distribution of Rothia dentocariosa in oral cavities. Journal of microbiological methods 91(1), 205-207.
- ↑ Schiff, M. J., & Kaplan, M. H. (1987) Rothia dentocariosa pneumonia in an immunocompromised patient. Lung 165(1), 279-282.
- ↑ Cell division protein FtsZ [Rothia dentocariosa]
- ↑ Ribonucleotide-diphosphate reductase subunit beta [Rothia dentocariosa]
- ↑ Zhou, Y., Yang, J., Zhang, L., Zhou, X., Cisar, J. O., & Palmer, R. J. (2016) Differential utilization of basic proline-rich glycoproteins during growth of oral bacteria in saliva. Applied and Environmental Microbiology 82(17), 5249-5258.
- ↑ Khan, S. T., Ahamed, M., Musarrat, J., & Al‐Khedhairy, A. A. (2014) Anti‐biofilm and antibacterial activities of zinc oxide nanoparticles against the oral opportunistic pathogens Rothia dentocariosa and Rothia mucilaginosa. European journal of oral sciences 122(6), 397-403.
This page is written by Alexandria Chin for the MICR3004 course, Semester 2, 2016
