Vibrio splendidus
Classification
Lineage:Bacteria; Proteobacteria; Gammaproteobacteria; Vibrionales; Vibrionaceae; Vibrio; Vibrio splendidus; Vibrio splendidus.
Species
Vibrio splendidus
Genome Structure
The genome of Vibrio splendidus consists of two circular chromosomes. The larger of the two, chromosome 1, is 3299 kb, and it contains 199 out of 205 of the minimal gene set. Chromosome 2 is 1675 kb long and is subject to more frequent genome rearrangements than chromosome 1, in which there is a higher conservation of gene order. Although differing in size, chromosomes 1 and 2 have similar G+C contents (44.03% G+C and 43.64% G+C, respectively) [7].
The genome of V. splendidus also contains 4498 predicted open reading frames, with 2998 on chromosome 1 and 1500 on chromosome 2. Chromosome 1 contains 7 rRNA genes and 97 tRNA genes, while chromosome 2 contains 1 rRNA gene and 17 tRNA genes [7].
Few mobile elements exist in the genome of V. splendidus. Chromosome 1 houses one RS1-like phage and 8 transposons, and chromosome 2 contains one transposon as well. Because all DNA repair enzymes are present in the organism, the high genotypic diversity between strains of V. splendidus is most likely not to be the result of a missing repair pathway. Rather, because it contains several genes that play a role in transformation competence (including those coding for a type IV pilus assembly complex and a putative DNA binding protein), it has been suggested that natural transformation may be one of the sources of high genomic polymorphism [7].
One surprising finding regarding the organism's genome is the nature of its chromosomal integron; in some strains of Vibrio splendidus, it is very small, while in others, it is completely absent from the genome. Prior to this finding, the chromosomal integron was thought to be a universal feature of all species in the Vibrio genus [7].
Cell Structure and Metabolism
Vibrio splendidus is a gram negative bacterium. It is rod-shaped, and has perichous polar flagella with sheaths. It is also bioluminescent, and for optimal growth it requires a salt concentration between 300 and 400 mM Na+ [9].
V. splendidus is chemoorganotrophic in nature, obtaining its energy and carbon from organic compounds. Its metabolism is facultatively fermentative; it makes ATP through aerobic respiration when oxygen is present, but it can also switch to fermentation when not in the presence of oxygen [8].
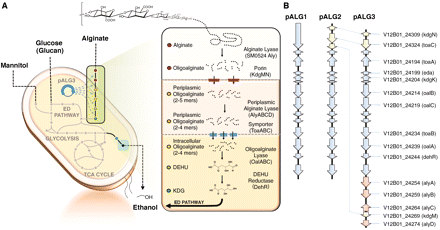
V. splendidus is also capable of metabolizing alginate, a linear block copolymer of uronic acids b-D-mannuronate (M) and a-L-guluronate (G). It contains a class of enzymes known as alginate lyases (Alys) which catalyze the depolymerization of alginate into oligomers through an endolytic b-elimination reaction. These oligomers can then be broken down further into unsaturated monomers by the exolytic enzyme oligoalginate lyase (Oal). The monomers produced here spontaneously rearrange into 4-deoxy-L-erythro-5-hexoseulose uronic acid (DEH), which is reduced into 2-keto-3-deoxygluconate (KDG) by DEH reductase (DehR). Using KDG kinase (KdgK) and KDG-6-phosphate aldolase (Eda), KDG eventually yields pyruvate and glyceraldehyde-3-phosphate [4].
Ecology and Pathogenesis
- Habitat
Vibrio splendidus is a dominant culturable Vibrio found in costal marine sediments, seawater and bivalve organisms [3].
- Ecology
Vibrio splendidus a planktonic vibrio species found in oceanic environment. It is a facultative anaerobe that has strains that have been found to be culturable in a lab environment. It has been found to be extremely prevalent in the gulf coast where it is able to be sampled out of most open water in the gulf.[11] V. splendidus strains have been found that emit a sort of bioluminescence that is visible in high cell concentration areas. A study has found that V. splendidus and Vibrio harveyi are so closely related that a site specific probe for V. harveyi was also found to hybridize with V. splendidus, although the true phylogenetic relationship between these two organisms is still unknown.[11] Vibrio Spendidus is a bioluminating organism. This means that the light it emits is not actual emitting a lot of thermal radiation.[12] It is commonly accepted that nearly 90% of marine animals emit some sort of bioluminescence, so just that fact could show how common this organism may be in aquatic environments. This light that is emitted by the organism can offer many benefits to any animal that chooses to uptake this microbe and associate it with itself.[12] It is speculated that the light could be used as an attractant luring prey in, as a deterrent keeping predators away or even simply a light source that allows the animal to see things that reflect the certain wavelength of light that V. splendidus is emitting. It is still not very well known what else this organism may be doing in marine life.[11]
- Pathology
The Vibrio splendidus' metallo protease gene Vsm is hypothesized to directly contribute to this organisms toxic effect on oysters . Other research is attempting to determine the toxic effects of this gene product on bivalves and to further determine its pathogenic ability [3]. V. splendidus has also been found to cause vibriosis in marine life.[10] Vibriosis in one of the most common diseases that is contracted by marine life, usually effecting sea life found in shallow waters during mid to late summer months.[10] By ingesting marine life that has this disease, vibriosis can be spread to humans. When contracted by humans, vibriosis exhibits many of the same symptoms as food poisoning such as nausea, vomitting, abdominal cramps and chills. When trying to care for someone with suspected vibriosis, just ensure they get enough fluids and the diseases should subsides in 3-4 days. Although hospitalization is rare, it can occur do to severe dehydration.
Significance and Current Research
Vibrio splendidus contains a 30 kilo-base pair fragment which codes for the molecular machinery needed for the transport and metabolism of alginate [4]. These cellular processes have the potential to greatly contribute to the production of biofuel from brown macroalgae [1].
Common substrates used for the production of biofuels, such as corn and sugar, present issues regarding production efficiency and competition with the production of food [4]. Current microbial technological research is being performed to address the cost and metabolic requirements to make use of the sugars released from recalcitrant polysaccharides found in other substrates such as brown macroalgae [5].
Brown macroalgae does not compete with food crops for the use of landmass, fertilizer or fresh water [4]. It is estimated that 59 dry metric tons/ha of this substrate can be produced per year and the estimated bio-ethanol activity from this crop is 19,000 liters/ha/year [4]. This ethanol yield is two-times greater than that of sugar and five-times than that of corn [4].
The most abundant sugars in brown macroalgae are alginate, mannitol and glucan [4]. An efficient method for the breakdown and metabolism of aliganate for the production of ethanol would allow brown macroalgae to be used at its full potential as a substrate for the production of biofuel.
The alginate transport and metabolizing pathway found in V. splendidus was reconstructed by creating a fosmid library using fragments of the Vibrio splendidus genomic DNA coding for the desired pathway and transforming it into E. coli [4]. It was reported that this engineered organism could produce ethanol at levels over 80% of the estimated maximum yield [4].
References
[1] Somerville C, Youngs H, Taylor C, Davis SC, Long SP. “Feedstocks for lignocellulosic biofuels”.“Science”. 2010 329(5993):790-2.
[2] Gay M, Berthe FC, Le Roux F. “Screening of Vibrio isolates to develop an experimental infection model in the Pacific oyster Crassostrea gigas.” “Diseases of Aquatic Organisms”. 2004 Apr 21;59(1):49-56.
[3] Le Roux F, Binesse J, Saulnier D, Mazel D. “Construction of a Vibrio splendidus mutant lacking the metalloprotease gene vsm by use of a novel counterselectable suicide vector.””Applied Environmental Microbiology”.2007 Feb;73(3):777-84.
[4] Wargacki AJ, Leonard E, Win MN, Regitsky DD, Santos CN, Kim PB, Cooper SR, Raisner RM, Herman A, Sivitz AB, Lakshmanaswamy A, Kashiyama Y, Baker D, Yoshikuni Y. “An engineered microbial platform for direct biofuel production from brown macroalgae.”“Science”. 2012 Jan 20;335(6066):308-13.
[5] Stephanopoulos G. “Challenges in engineering microbes for biofuels production.”“Science”. 2007 Feb 9;315(5813):801-4.
[6] Shin Y, Liu J, Wang LQ, Nie Z, Samuels WD, Fryxell GE, Exarhos GJ. “Ordered Hierarchical Porous Materials: Towards Tunable Size- and Shape-Selective Microcavities in Nanoporous Channels The Pacific Northwest National Laboratory is operated by Battelle for the U.S. Department of Energy under Contract DE-AC06-76RL0 1830. This work is supported by the Office of Basic Energy Sciences, Division of Materials Sciences, of the U.S. Department of Energy.” “Angewandte Chemie International Edition”. 2000 Aug 4;39(15):2702-2707.
[7]Le Roux F, Zouine M, Chakroun N, Binesse J, Saulnier D, Bouchier C, Zidane N, Ma L, Rusniok C, Lajus A, Buchrieser C, Médigue C, Polz MF, Mazel D. "Genome sequence of Vibrio splendidus: sn abundant planctonic marine species with a large genotypic diversity." Environmental Microbiology. 2009 Aug;11(8):1959-70.
[8] Thompson FL, Gevers D, Thompson CC, Dawyndt P, Naser S, Hoste B, Munn CB, Swings J. "Phylogeny and Molecular Identification of Vibrios on the Basis of Multilocus Sequence Analysis." Applied Environmental Microbiology. 2005 Sep;71(9):5107-15.
[9] Baumann P, Baumann L, Bang S, Woolkalis M. "Reevaluation of the Taxonomy of Vibrio, Beneckea, and Photobacterium: Abolition of the Genus Beneckea." Current Microbiology. 1980;4:127-132.
[10] Jensen S, Samuelson O, Anderson K, Torkildsen L, Lambert C, Choquet C,Paillard C, Bergh O. "Characterization of strains of Vibrio splendidus and V. tapetis isolated from corkwing wrasse Symphodus melops suffering vibriosis" Diseases of Aquatic Organisms. 53: 25–31, 2003
[11] Mealson K.H., Wimpee C, Wimpee B. "Identification of Vibrio splendidus as a Member of the Planktonic Luminous Bacteria from the Persian Gulf and Kuwait Region with lux,4 Probes" APPLIED AND ENVIRONMENTAL MICROBIOLOGY. 59:2684-2689, 1993
[12] Cobbold C, Rink T. "Fluorescence and bioluminescence measurement of cytoplasmic free calcium" Biochem J. 248(2): 313–328
Author
Page authored by Laura Carroll, RonJon Casanova-Smith and Ashlee Bruewer, students of Prof. Jay Lennon at Michigan State University.
<-- Do not remove this line-->