Zinc Finger Nuclease (ZFN): Difference between revisions
| Line 35: | Line 35: | ||
[[Image:knockout.png|thumb|300px|right|Figure 2.]] | [[Image:knockout.png|thumb|300px|right|Figure 2.]] | ||
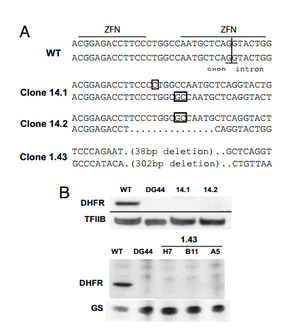
Santiago et. al tested the ability of ZFNs to create knockout mammalian cells. To create knockouts, the same principles described in the introduction were followed. That is, nonhomologous end joining imperfectly repairs the DNA after a double stranded break, leading to nonsense frame-shift mutations. The group knocked out the dihydrofolate reductase (DHFR) gene in the ovary cell line of a Chinese hamster. | |||
A ZFN-based approach to knockout generation was found to be very feasible. The one-step technique much faster than other conventional methods. Double stranded breaks in DNA increase the frequency of homologous recombination by more than 1000. Thus, a high-frequency (2-3%) of mutants were reported after ZFN treatment. The knockout mutants could then be isolated out and cloned separately. When using methods where mutations are less frequent, the creation of knockout genes must be coupled with a selection method that selects for the low frequency mutants and fleshes them out. Because ZFNs directly affect the genome, gene knockouts are permanent and conserved in new generations. | |||
<br> [http://www.pnas.org/content/105/15/5809.short <sup>1</sup>]<br> | <br> [http://www.pnas.org/content/105/15/5809.short <sup>1</sup>]<br> | ||
Revision as of 03:59, 6 December 2013
Introduction

Zinc finger nucleases (ZFNs) are proteins composed of a DNA binding-domain and a DNA cleaving-domain. One zinc “finger” binding domain recognizes and binds to a three nucleotide sequence, one can thus increase the binding specificity of a ZFN by adding more zinc fingers. The DNA cleaving-domain (Fok 1 nuclease) can then be used to create a double-stranded break in DNA at the desired point (Fig. 1).
After a double stranded break, DNA is repaired by non-homologous end joining. This often causes small insertions or deletions in the DNA, resulting frame-shift mutations. These frame-shifts result in nonsense mutations or nonsense-mediated decay. These effects are therefore very useful for creating knockout genes.
Additionally, genetic material such as DNA plasmids can be integrated into a gene after a double stranded break. A desired donor plasmid will contain arms of DNA which are homologous to the ZFN cut site. The donor plasmid and ZFNs can then be added simultaneously to the cell, the genetic material will be successfully incorporated after a double stranded cut and subsequent homologous recombination. Though homologous recombination is a rare event in most cells, double stranded breaks from ZFNs greatly increase the frequency of such events, facilitating gene insertion.
This makes ZFNs very useful tools for creating transgenic organisms and genetic engineering.
History
While other gene targeting procedures have existed for quite some time, the success of earlier methods were dependent on several factors including homologous recombination. Also, while genetic changes were observed in fungi like Saccharomyces cerevisiae and bacteria, most methods did not work on eukaryotic organisms. However, when studies in the 80s revealed that double-stranded DNA breaks greatly increased the frequency of recombination and thus successful genetic modifiactions, tools such as ZFNs became favored. Previously, stem cells were required (Rudin et. al.).
The insertion code consists of:
Double brackets: [[
Filename: PHIL_1181_lores.jpg
Thumbnail status: |thumb|
Pixel size: |300px|
Placement on page: |right|
Legend/credit: Diagram of a zinc finger nuclease. The zinc fingers are part of the binding-domain and have precisely identified and bound to a group of matching nucleobases in the DNA sequence. The cleaving-domains are Fok 1, which make a break in the DNA in both strands. Image credit to Stewart, C. N. and Burris, J. Jr.
Closed double brackets: ]]
Other examples:
Bold
Italic
Subscript: H2O
Superscript: Fe3+
Using ZFNs to create knockout cells
Santiago et. al tested the ability of ZFNs to create knockout mammalian cells. To create knockouts, the same principles described in the introduction were followed. That is, nonhomologous end joining imperfectly repairs the DNA after a double stranded break, leading to nonsense frame-shift mutations. The group knocked out the dihydrofolate reductase (DHFR) gene in the ovary cell line of a Chinese hamster.
A ZFN-based approach to knockout generation was found to be very feasible. The one-step technique much faster than other conventional methods. Double stranded breaks in DNA increase the frequency of homologous recombination by more than 1000. Thus, a high-frequency (2-3%) of mutants were reported after ZFN treatment. The knockout mutants could then be isolated out and cloned separately. When using methods where mutations are less frequent, the creation of knockout genes must be coupled with a selection method that selects for the low frequency mutants and fleshes them out. Because ZFNs directly affect the genome, gene knockouts are permanent and conserved in new generations.
1
Using ZFNs to insert genes into an existing genomic site
Include some current research in each topic, with at least one figure showing data.
TALENs
Include some current research in each topic, with at least one figure showing data.
Conclusion
Overall paper length should be 3,000 words, with at least 3 figures.