Agrobacterium tumefaciens and genetic engineering
Introductionn

By George Ni
Genetic engineering is the manipulation of DNA sequences in organisms using biotechnology. This process involves knocking out DNA sequences from organisms, as well as changing and adding new segments of DNA. Genetic engineering is widely used in biology research, food production, and medical therapies to alter phenotypes and functions of target organisms for desired traits. [1] Agrobacterium tumefaciens is a rod-shaped and gram-negative soil bacterium. This species of bacteria infects plants by inducing crown gall disease. A.tumefaciens could infect the healthy plants with a small segment of DNA from the plasmids of bacteria called T-DNA. The infected DNA segment is tumor-inducing. It triggers hormone release by the plant's cells and stimulates tumorous growth. Tumor cells in plants then release “opines”, which can be used by A.tumefaciens as a source of carbons and nitrogen for metabolism. [2]
The ability of A.tumefaciens to integrate DNA segments into plant cells is widely used by scientists in the field of genetic engineering. In the engineered A.tumefaciens, the oncogene in the plasmids is knocked out and replaced by the gene of interest. Without destroying the ability for A.tumefaciens to integrate its DNA into a plant cell, the engineered plasmids successfully induce the gene of interest such as traits that promote crops growth and make the fruits taste better. [2]
Agrobacterium-mediated transformation
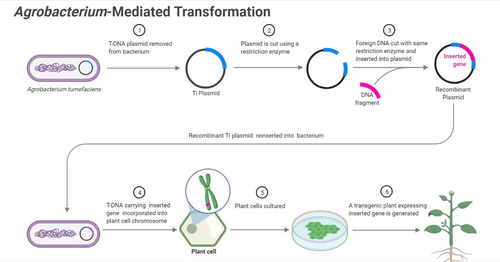
Agrobacterium genus is well-known for its ability to transfer genetic material, specifically DNA, into plant cells, which has made it an important tool for the production of genetically modified (GM) crops. Agrobacterium tumefaciens is the most commonly studied species for genetic engineering. [3]
Researchers have harnessed the ability of Agrobacterium. to introduce target DNA segments into plant cells for genetic engineering purposes. The process of utilizing T-DNA of Agrobacterium genus is called Agrobacterium-mediated transformation (AMT). This process requires researchers to expose the engineered Agrobacterium cells to plant tissues. After Agrobacterium successfully transfers T-DNA to the plants, the target DNA segments will be integrated into plant cells, and the target genes can be further replicated by plant cells. .[4] A.tumefaciens is most frequently used species to conduct AMT because of its relatively simple genetic makeup and well-characterized mechanisms for T-DNA transfer of DNA into plants.
In addition, the research team led by Michielse have found that the efficiency of AMT increases when several genes of Wild Type A.tumefaciens are mutated. [4] They first discovered that mutations in A.tumefaciens genes chvA, chvB, or exoC don’t affect the efficiency of AMT. These genes are responsible for the production of synthesis and export of β-1,2-glucan, as well as the attachment and subsequent transformation in the plant infection process. However, the mutated versions of chvA, chvB, or exoC don’t affect AMT. The roles of these genes in AMT are rejected by Michielse and his research team. [4] Besides, they found that the inactivation of gene Cel1, which is responsible for the cellulose production, has led to a seven-fold increase in AMT efficiency. Their result showed that the cellulose fibers have a negative influence on the attachment and infection of A.tumefaciens to their hosts. [4] The discovery led by Michielse and his research team has helped the scientists to design specific mutated A.tumefaciens strain to conduct AMT in a much more efficient way.
Genome Structure
The unique action mode of A.tumefaciens benefits from its genomic structure. A.tumefaciens contains a mix of linear and circular chromosomes. [5] Its circular plasmids promote the invasion of genetic material to its host. Circular plasmids are smaller in size compared to the chromosome of bacteria. While genes in chromosomes have instructions for basic functions of bacterial cells, such as cell division metabolis, genes in plasmids usually code for secondary functions, such as antibiotics resistance and cell to cell communication. [6] Therefore, A.tumefaciens utilized the characteristics that plasmids are small, and the transmission of plasmids genes are not lethal to achieve infections. T-DNA is the segment of DNA that is excised from the Ti-plasmid of A.tumefaciens that is transferred into the target plants. T-DNA in plasmids is transferred and incorporated into the host plants’ genome when the infection process is completed.
Two component regulators
Moreover, the infection process requires two component regulators of virA and virG in the vir operon by A.tumefaciens cells. The wounded plant tissue is the environmental signal to trigger A.tumefaciens cells to bind. A.tumefaciens cells could make complex biofilms at the colonization site on wounded tissues. [7] The monosaccharides of sugars released by plants are detected by virA transmembrane receptor kinase. The receptor kinase then triggers autophosphorylation. Phosphate is transferred to virG, which is the activator that initiates the transcription of vir operon. The proteins generated by vir operon facilitate the transportation of T-DNA into the plant cells. T-DNA is then integrated into the plant genomes by non-homologous recombination. [8]
In addition, virA and virG two component systems are significant mediators of signal transduction that enables A.tumefaciens to respond quickly to the environmental change. Recent evidence has shown that virA and virG two component systems can adapt to function in tobacco protoplasts. The vir operon can be efficiently transcribed with eukaryotic transcriptional apparatus in tobacco protoplasts. [9] The system allows the plants to react quickly to environmental changes such as pH.
Genes in T-DNA
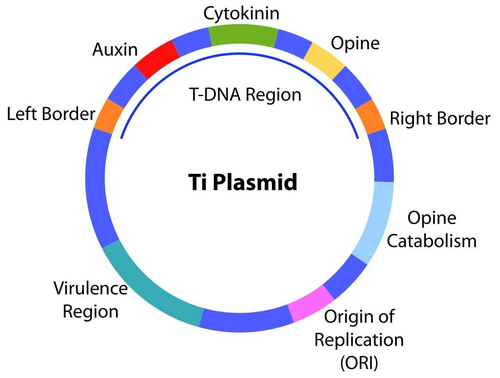
Genes in T-DNA of A.tumefaciens are mainly involved in transferring genetic materials into plants, extracting nutrients from the plants and resulting in formation of tumors in the plants. Three components of T-DNA helping A.tumefaciens to achieve these functions are border sequences, transfer genes and oncogenes. [10] Border sequences are the recognition sites for the excision of T-DNA from the Ti-plasmid. The border sequence is significant in helping A.tumefaciens to excise all sequences of T-DNA, while not cutting other parts of the plasmids. Transfer genes are mainly responsible for the transfer process of T-DNA into the plant cells. Transfer genes include virA, virD1, virD2, virD3, VirF and virG. After Vir A detects the sugar released by plants, Vir G is auto phosphorylated, which triggers increased Vir operon expression. [10] As a result, virD1, virD2, virD3, VirF are produced. Vir D1 and Vir D2 are involved in the protection of T-DNA and integration of T-DNA into plants' genomes. VirD3 and Vir F play an important role in the transfer process of T-DNA..[10] Oncogenes of T-DNA directly result in the uncontrolled cell growth and tumor formation in the plants. The oncogenes could trigger the synthesis of many growth hormones by the infected plants, such as the release of auxins and cytokinin. [11] The normal developmental cycle of plants is disrupted, and tumors can form as a result of uncontrolled development.
Researchers have identified some oncogenes. Two A.tumefaciens T-DNA auxin biosynthesis genes products called iaaM and iaaH are identified to trigger the auxin overproduction and uncontrolled cell growth in plants. [12] These two genes work together to encode enzymes that produce indoleacetic acid (IAA), which is a common plant hormone of auxin class. In particular, iaaM encodes tryptophan monooxygenase to convert tryptophan to indoleacetamide (IAM). iaaH then encodes indoleacetamide hydrolase to convert IAM to IAA. [12] The release of IAA promotes into plant tissues causes cell elongation and cell growth in the plants. Auxin is essential for the regulation of plant growth and development. Most plants are very sensitive to the change of Auxin. Changes in micromolar amounts of auxins can cause dramatic changes in plant phenotypes and development, while changes in millimolar amounts of auxins can be lethal to plants. [12] This also explains why infection by A.tumefaciens induces tumor formation in plants. Severe infections by A.tumefaciens to susceptible plant species can even lead to plant death.
Conjugation in A.tumefaciens
Besides transferring genetic materials to the plant cells, A.tumefaciens can also transfer its genetic materials to other bacteria by conjugation. Conjugation is a type of horizontal gene transfer, which is the transfer of genetic material between bacteria that are not parent and offspring. [13] During conjugation, A.tumefacienss can transfer the Ti plasmid to other bacteria in close proximity through a specialized structure called a conjugative pilus, which forms a physical bridge between the donor bacterium and the recipient bacterium. [13] The Ti plasmid is then transferred from the donor to the recipient, and the recipient bacterium can acquire the ability to cause crown gall disease in plants if it receives the complete Ti plasmid, including the T-DNA region. The mechanism of conjugation allows A.tumefaciens to spread its virulence genes to other A.tumefaciens cells. The recipient cells can then gain the ability to infect crown gall disease to different plant tissues.
Conjugation is also a common genetic transfer mechanism found in other bacterial species. This could even threaten the human healthcare system. When a bacterium that carries antibiotic resistance genes conjugates with another bacterium that lacks those resistance genes, it can transfer the genes conferring antibiotic resistance to the recipient bacterium. This can contribute to the rapid spread of antibiotic resistance bacteria in diverse environments, such as hospitals, malls and social communities. The transfer of antibiotic resistance genes through conjugation can greatly impact the effectiveness of antibiotics in treating bacterial infections. It can lead to the spread of antibiotic resistance among bacterial populations, making it more difficult to treat infections and diseases with commonly used antibiotics.
There are many examples of antibiotic resistance genes that can be transferred among bacteria through conjugation, leading to the spread of antibiotic resistance. Extended-spectrum beta-lactamase (ESBL) genes encode enzymes that can inactivate a broad range of beta-lactam antibiotics, including penicillin, cephalosporins, and monobactams. [14] ESBL genes are often carried on plasmids, which can be transferred through conjugation. ESBL genes can even transfer among different bacterial species. It allows many bacterial species to acquire resistance to multiple antibiotics simultaneously. Moreover, Carbapenems are a class of antibiotics that are considered as last-resort drugs for the treatment of serious infections caused by multidrug-resistant bacteria. However, bacteria that carry Carbapenemase genes can inactivate Carbapenem antibiotics, leading to Carbapenem resistance. [15] Carbapenemase genes can be transferred through conjugation, leading to the spread of Carbapenem resistance among different bacterial species. Carbapenems resistance can be lethal to patients, but the current medical treatments have minimal effects in killing bacteria with Carbapenems genes.
Therefore, it can be seen that conjugation among bacteria can make many antibiotic drugs ineffective for treatments. It also stresses the importance of careful dosage of antibiotic use, infection control measures, and strategies to minimize the spread of antibiotic resistance. If conjugation among bacterial populations can be effectively blocked, it can solve many clinical issues raised by antibiotic resistant bacteria.
Genetic engineering in Tobacco plants

A.tumefaciens has been widely used as a vector to introduce desirable traits into tobacco plants through genetic engineering. Collagen I have been used in various medical treatments and applications due to its unique properties. For example, it has been used in wound dressings and scaffolds for tissue engineering to promote wound healing. Collagen-based dressings provide a protective barrier while also providing a favorable environment for cell migration, proliferation, and tissue regeneration. [16] Collagen I is usually extracted from animal tissues. However, infections and other contaminants happen when this material is extracted for medical use. Many pharmaceutical industries are particularly interested in finding the alternative source for Collagen I. C Merle and her research team showed that Agrobacterium tumefaciens-mediated transgenic tobacco plants are great candidates for massive production of Collagen I. [17]
In M Merle and her colleagues’ previous research, they have shown that tobacco plants can produce recombinant collagen (rColl I) as a fully processed triple helix. However, the thermal stability of the triple helix is below the standard for the medical use. The analysis of the chemical structure of rColl I indicates that hydroxyproline residues are needed to stabilize the overall structure of rColl I. Even though Tobacco plants have their own proline-4-hydroxylases (P4Hs), they are unable to insert the hydroxyproline into the amino acid chains of rColl I. Researchers have found that P4Hs from Caenorhabditis elegans (C. elegans) can combine well with rColl I in tobacco plants. In addition, P4Hs from Caenorhabditis elegans can also function even in a low temperature environment. It largely increases the thermal stability of rColl I in tobacco plants. [17]
The researchers used A.tumefaciens as the vector to transfer genes that produce P4Hs from C.elegans into tobacco plants. [17] The transient assay of A.tumefaciens is cost-effective and suitable for usage in large-scale production. Vacuum infiltration of tobacco plants with A.tumefaciens strains A1286 (containing C. elegans P4H α subunit) has been processed for four days. Purified Collagen I is analyzed by 6% SDS–PAGE. rColl I from the transformed tobacco plants contain similar molecular weight compared to the control rColl I. In addition, analysis of the amino acid compositions in transformed tobacco plants reveals that the presence of hydroxyproline in rColl I is 6.85%, while the hydroxyproline in rColl I of untransformed tobacco plants is only 0.68%. Hydroxyl proline residues are successfully inserted into the rColl I for Agrobacterium tumefaciens-mediated transgenic tobacco plants. Finally, the thermal stability measurement indicates that rColl I for transformed tobacco plants is resistant to pepsin up to 36.8 °C, which is significantly increased compared to the untransformed plants. As a result, the result shows that rColl I produced by Agrobacterium tumefaciens-mediated transgenic tobacco plants is qualified for medical use.
Genetic engineering in crop improvement
In addition to medical use, A.tumefaciens genetic engineering has also been utilized as a powerful tool for crop improvement. By exploiting its natural ability to transfer DNA into the genomes of plants, A.tumefaciens has been employed to introduce desirable traits into crops, leading to improved agricultural production. [18] For instance, genes that confer resistance to pests, diseases, or environmental stress have been successfully inserted into crops, providing increased yield and reduced crop losses. Additionally, A.tumefaciens has been used to develop herbicide-tolerant crops, enabling more efficient weed control. [18] These advancements in crop improvement using A.tumefaciens have the potential to contribute to sustainable agriculture by enhancing crop productivity and reducing the reliance on chemical pesticides.
Usually, selectable marker genes are integrated into the T-DNA of A.tumefaciens. Different crop species need different marker genes to increase its viability and production in the environment. Researchers carefully select the corresponding marker genes based on the crop species. For example, the most commonly used markers in cereals to confer resistance to antibiotics and herbicides hygromycin phosphotransferase (hpt) and phosphinothricin acetyltransferase (pat). Gene hpt is the best selectable marker for rice and barley, while gene pat is most selectable for maize. [19]
Furthermore, most marker genes are tissue-specific, and T-DNA of A.tumefaciens must be transferred to the specific tissue type to induce the gene expression. For example, rice globulin (Glb) promoter and wheat glutenin gene promoter (Glu) are used for endosperm-specific gene expression. Chlorophyll a/b binding (cab) gene is leaf-specific for rice. [19] The tissue-specific induction of gene expression is also seen in other techniques of genetic engineering, such as promoter engineering. [20]
When transferring T-DNA of A.tumefaciens into crops, several key factors should be considered, which include type of stage of the plants, the strain of Agrobacterium, type of vectors, acetosyringone and co-cultivation temperature. According to the previous experiments, plant tissues in active dividing stages are most suitable for the transformation since there will be higher frequencies of expression for the incorporated genes. For Agrobacterium strains, it has been shown that the super-virulent strain is most suitable for the plant transformation. Strains including LBA4404, EHA101 and EHA105 contain super-binary vectors that are more efficient for the integration of T-DNA into the plant tissues. [19]
Conclusion
In conclusion, A.tumefaciens has revolutionized genetic engineering and biotechnology by serving as a powerful tool for introducing foreign genes into plants. Its natural ability to transfer DNA to plant cells has been harnessed to create genetically modified organisms (GMOs) with desired traits, such as resistance to pests, and tolerance to environmental conditions. This has led to significant advancements in agriculture, including increased crop yields, and improved food quality. Genetic engineering of A.tumefaciens is also utilized to design medicines with higher tolerance to temperature variation and increase the cost-performance ratio for pharmaceutical industries.
However, the use of A.tumefaciens in genetic engineering also raises ethical, social, and environmental concerns. Critics argue that GMOs may have unknown long-term effects on ecosystems and human health and may contribute to the loss of biodiversity and the dependence on large seed corporations. [21] Therefore, it is crucial to conduct thorough risk assessments and adopt responsible practices when using A.tumefaciens and other genetic engineering techniques.
Despite the controversies surrounding GMOs, A.tumefaciens remains a valuable tool in plant biotechnology, with immense potential for addressing global challenges such as food security, health issues, and sustainable agriculture. Continued research and regulatory oversight are essential to ensure the safe and responsible use of A.tumefaciens and other genetic engineering technologies, while balancing the benefits and risks associated with GMOs. Overall, A.tumefaciens has paved the way for genetic engineering to become a powerful tool in modern agriculture, offering a promising future for crop improvement and medical treatments.
References
[1]Paoletti, M. G., & Pimentel, D. (1996). Genetic Engineering in Agriculture and the Environment. BioScience, 46(9), 665–673. https://doi.org/10.2307/1312896
[2]Gohlke, J., & Deeken, R. (2014). Plant responses to Agrobacterium tumefaciens and crown gall development. Frontiers in Plant Science, 5. https://www.frontiersin.org/articles/10.3389/fpls.2014.00155
[3]Paoletti, M. G., & Pimentel, D. (1996). Genetic Engineering in Agriculture and the Environment. BioScience, 46(9), 665–673. https://doi.org/10.2307/1312896
[4]Michielse, C. B., Hooykaas, P. J. J., van den Hondel, C. A. M. J. J., & Ram, A. F. J. (2005). Agrobacterium-mediated transformation as a tool for functional genomics in fungi. Current Genetics, 48(1), 1–17. https://doi.org/10.1007/s00294-005-0578-0
[5]Wood, D. W., Setubal, J. C., Kaul, R., Monks, D. E., Kitajima, J. P., Okura, V. K., Zhou, Y., Chen, L., Wood, G. E., Almeida, N. F., Woo, L., Chen, Y., Paulsen, I. T., Eisen, J. A., Karp, P. D., Bovee, D., Chapman, P., Clendenning, J., Deatherage, G., … Nester, E. W. (2001). The Genome of the Natural Genetic Engineer Agrobacterium tumefaciens C58. Science, 294(5550), 2317–2323. https://doi.org/10.1126/science.1066804
[6]Bouma, J. E., & Lenski, R. E. (1988). Evolution of a bacteria/plasmid association. Nature, 335(6188), 351–352. https://doi.org/10.1038/335351a0
[7]Tomlinson, A. D., & Fuqua, C. (2009). Mechanisms and regulation of polar surface attachment in Agrobacterium tumefaciens. Current Opinion in Microbiology, 12(6), 708–714. https://doi.org/10.1016/j.mib.2009.09.014
[8]Gao, R., & Lynn, D. G. (2005). Environmental pH Sensing: Resolving the VirA/VirG Two-Component System Inputs for Agrobacterium Pathogenesis. Journal of Bacteriology, 187(6), 2182–2189. https://doi.org/10.1128/JB.187.6.2182-2189.2005
[9]Czarnecka-Verner, E., Salem, T. A., & Gurley, W. B. (2016). Adaptation of the Agrobacterium tumefaciens VirG response regulator to activate transcription in plants. Plant Molecular Biology, 90(3), 217–231. https://doi.org/10.1007/s11103-015-0407-x
[10]Jin, S., Roitsch, T., Ankenbauer, R. G., Gordon, M. P., & Nester, E. W. (1990). The VirA protein of Agrobacterium tumefaciens is autophosphorylated and is essential for vir gene regulation. Journal of Bacteriology, 172(2), 525–530. https://doi.org/10.1128/jb.172.2.525-530.1990
[11]Akiyoshi, D. E., Klee, H., Amasino, R. M., Nester, E. W., & Gordon, M. P. (1984). T-DNA of Agrobacterium tumefaciens encodes an enzyme of cytokinin biosynthesis. Proceedings of the National Academy of Sciences, 81(19), 5994–5998. https://doi.org/10.1073/pnas.81.19.5994
[12]The effects of overproduction of two Agrobacterium tumefaciens T-DNA auxin biosynthetic gene products in transgenic petunia plants. (1987). Genes & Development, 1(1), 86–96. https://doi.org/10.1101/gad.1.1.86
[13]Waters, V. L. (2001). Conjugation between bacterial and mammalian cells. Nature Genetics, 29(4), 375–376. https://doi.org/10.1038/ng779
[14]Salinas, L., Loayza, F., C, árdenas P., Saraiva, C., Johnson, T. J., Amato, H., Graham, J. P., & Trueba, G. (2021). Environmental Spread of Extended Spectrum Beta-Lactamase (ESBL) Producing Escherichia coli and ESBL Genes among Children and Domestic Animals in Ecuador. Environmental Health Perspectives, 129(2), 027007. https://doi.org/10.1289/EHP7729
[15]Yoon, E.-J., & Jeong, S. H. (2021). Mobile Carbapenemase Genes in Pseudomonas aeruginosa. Frontiers in Microbiology, 12. https://www.frontiersin.org/articles/10.3389/fmicb.2021.614058
[16]Shoulders, M. D., & Raines, R. T. (2009). Collagen Structure and Stability. Annual Review of Biochemistry, 78(1), 929–958. https://doi.org/10.1146/annurev.biochem.77.032207.120833
[17]Merle, C., Perret, S., Lacour, T., Jonval, V., Hudaverdian, S., Garrone, R., Ruggiero, F., & Theisen, M. (2002). Hydroxylated human homotrimeric collagen I in Agrobacterium tumefaciens-mediated transient expression and in transgenic tobacco plant. FEBS Letters, 515(1–3), 114–118. https://doi.org/10.1016/S0014-5793(02)02452-3
[18]Van Wordragen, M. F., & Dons, H. J. M. (1992). Agrobacterium tumefaciens-mediated transformation of recalcitrant crops. Plant Molecular Biology Reporter, 10(1), 12–36. https://doi.org/10.1007/BF02669262
[19]Singh, R. K., & Prasad, M. (2016). Advances in Agrobacterium tumefaciens-mediated genetic transformation of graminaceous crops. Protoplasma, 253(3), 691–707. https://doi.org/10.1007/s00709-015-0905-3
[20]Blazeck, J., & Alper, H. S. (2013). Promoter engineering: Recent advances in controlling transcription at the most fundamental level. Biotechnology Journal, 8(1), 46–58. https://doi.org/10.1002/biot.201200120
[21]Kangmennaang, J., Osei, L., Armah, F. A., & Luginaah, I. (2016). Genetically modified organisms and the age of (Un) reason? A critical examination of the rhetoric in the GMO public policy debates in Ghana. Futures, 83, 37–49. https://doi.org/10.1016/j.futures.2016.03.002
