Caulobacter crescentus in Biotechnology
Overview
Caulobacter crescentus is a non-pathogenic, aquatic, Gram-negative bacterium.[9] One of its most distinguishing characteristics is the surface layer (S-layer) of its outermost cell wall component.[1] The S-layer is composed a single protein, RsaA, and is among the most abundant proteins in C. crescentus, accounting for 31% of total cell protein, and up to 51% when multiple copies of rsaA are present. This is the highest observed level of RsaA protein expression than any other protein in any bacterium. [5]
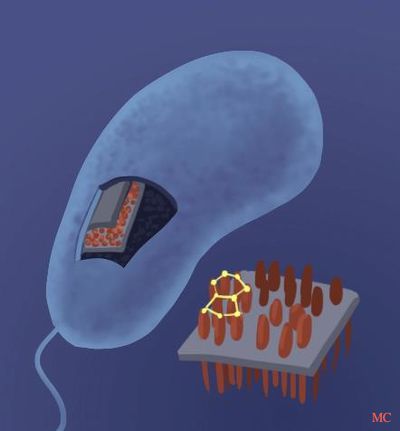
Significance
Due to the non-pathogenic nature of C. crescentus [3] and its high expression of RsaA protein, it has been used to express and assemble foreign proteins at high concentrations.[6] C. crescentus is being used in a variety of biotechnological applications such as in cancer therapy, anti-HIV microbicide, whole cell vaccines, and cytokine display.[1]. All these employ recombinant DNA methods to create modified versions of the rsaA gene, which are then translated to fusion proteins. These methods generally involve the insertion of the desired gene into a specific insertion site. The premise is that through use of molecular cloning techniques, genetic material from multiple sources are brought together.
Other beneficial characteristics that C. crescentus possess include its ability to be stored for extended periods of time, and its relatively inexpensive production which can be applied to a large scale.[8] Surprisingly, wild type C. crescentus has also been shown to have immuno-stimulatory properties[5] and may also be used as a prophylactic agent.
Use in Biotechnology
Difficulties with Structure Analysis
The S-layer protein of C. crescentus has not yet been visualized by X-Ray crystallography. Current difficulties include limitations in resolving the organization and structure of the assembled S-layer at atomic resolutions. This is a consequence of the two-dimensional assembly of RsaA, preventing the proteins from being analyzed by conventional methods such as X-ray crystallography, which require three-dimensional crystals. Such high-resolution analysis is crucial to understand and therefore predict where proteins are displayed within the array of S-layer monomers.[1] Despite difficulties in obtaining the atomic resolution needed for the structural analyses described above, successful attempts have been made in secreting various recombinant S-layer proteins whose assembly and attachment are unimpeded.[5]
C. crescentus and Immuno-Stimulatory Properties
During initial cancer vaccine studies, unexpected results in the control group showed that wild type C. crescentus displayed anti-tumor properties in three different strains of mice, each challenged with a different transplantable tumor model. Both live and heat killed C. crescentus were used to vaccinate mice three times (two weeks between immunizations) by intraperitoneal injections. The mice were challenged with different tumors two weeks after the final immunization, and observed for three months.[3] In the Lewis lung carcinoma model, 60% of the mice pre-immunized with live C. crescentus were still alive by day 90, whereas all mice pre-immunized with heat killed C. crescentus or the saline control died by 8 weeks post tumor challenge. Similar results were obtained in the murine mammary carcinoma model.[3] In the murine L1210 leukemia model, the entire control group only survived until the fourth week, whereas approximately 80% of mice pre-immunized with live C. crescentus survived, and 100% of the heat-killed immunized mice survived the entire 3-month study period.[3] These findings lead to the conclusion that C. crescentus can function as a prophylactic cancer vaccine in three murine tumor models, attributable to the host immune response generated. This presents a case of bacteria exhibiting immuno-stimulatory properties.[3]
C. crescentus and HIV
The display capabilities of C. crescentus can be used to develop antibodies, which can mimic host receptors or co-receptors, to inhibit host infection. This is possible because the respective native binding sites of the expressed proteins remain intact. One great application is the introduction of domain 1 of CD4 or MIP1α to inhibit HIV infection. CD4 serves as the HIV receptor, and MIP1α as the ligand for CCR5 HIV co-receptor. Depending on the HIV-1 clones, 35-78% reduction in infection was found with MIP1α displaying C. crescentus (Cc-MIP1α), and 22-56% block in infection for CD4 displaying C. crescentus (Cc-CD4). There was no difference in effectiveness between heat inactivated or live Caulobacter on inhibition of HIV-1 infection.[6] The most effective inhibition of HIV-1 infection was shown to come from a cocktail of several recombinant Caulobacters, each expressing different infection blockers. This combinatorial effect was shown to inhibit infection from 74-98% depending on the reference strains of HIV-1. This enhancement in infection reduction shows that an effective microbicide would have to be a combinatorial mix to maximally inhibit HIV infection.[6] Recent research has shown enhanced neutralization of HIV when the antibodies are displayed on the surface of the S-layer, instead of secreted antibodies.[4,7]
C. crescentus and Pseudomonas aeruginosa vaccine candidate
The novel protein secretion system of C. crescentus was used to produce vaccine candidates against Pseudomonas aeruginosa. Caulobacter’s RsaA protein was adapted to make 2 recombinant proteins of the pilus epitope (termed adhesintopes) using the PE and PCK portions of the P. aeruginosa pilin. These fusion proteins were then used as a vaccine candidate in a mouse challenge model. The PCK fusion proteins were compared to previous adhesintope-based vaccine candidates against P. aeruginosa and the PCK fusion protein induced a 1000-fold greater antibody response, balanced in IgG1 and IgG2 antibody response. However, when these immunized mice were then challenged with a P. aeruginosa infection, none of the fusion proteins provided a significant level of protection.[10] Despite the lack of immunity afforded by the vaccine, the methods used present a new system of vaccine production that can be used to quickly produce vaccine proteins.[3,10]
References
1. Amat F., Comolli L.R., Nomellini J.F., Moussavi F., Downing K.H., Smit S., and Horwitz M. “Analysis of the Intact Surface Layer of C. crescentus by Cryo-Electron Tomography.” Journal of Bacteriology, 2010, DOI: 10.1128/JB.00747-10
2. Awram, P., Smit, J. “The Caulobacter crescentus paracrystalline S-layer protein is secreted by an ABC transporter (type I) secretion apparatus.” Journal of Bacteriology, 1998, DOI: 10.1128/JB.182.11.3298-3301.2000
3. Bhatnagar P.K., Awasthi A., Nomellini J.F., Smit J., and Suresh M.R. "Anti-Tumor Effects of the Bacterium C. crescentus in Murine Tumor Models.” Cancer Biology & Therapy, 2006, DOI: 10.4161/cbt.5.5.2553
4. Duval M., Lewis C.J, Nomellini J.F., Horwitz M.S., Smit J., Cavacini L.A. “Enhanced Neutralization of HIV by Antibodies Displayed on the S-Layer of Caulobacter crescentus.” American Society for Microbiology, 2011, DOI: 10.1128/AAC.00509-11
5. Lau J.H.Y, Nomellini J.F., and Smit J. “Analysis of high-level S-layer protein secretion in C. crescentus.” Canadian Journal of Microbiology, 2010, DOI: 10.1139/can.j.microbiol.10-036
6. Nomellini J.F., Li C., Lavallee D., Shanina I., Cavacini L.A., Horwitz M.S., and Smit J. “Development of an HIV-1 Specific Microbicide Using C. crescentus S-Layer Mediated Display of CD4 and MIP1α.” PLoS ONE, 2010, DOI: 10.1371/journal.pone.0010366
7. Nomellni J.F., Duncan G., Dorocicz I.R., Smit J. “S-Layer-Mediated Display of the Immunoglobulin G-Binding Domain of Streptococcal Protein G on the Surface of Caulobacter crescentus: Development of an Immunoactive Reagent.” Applied and Environmental Microbiology, 2007, DOI: 10.1128/AEM.02900-06
8. Smit J. “Heads Up S-Layer Display: The Power of Many.” Structure, 2008, DOI: 10.1016/j.str.2008.07.003
9. Umelo-Njaka E., Bingle W.H., Borchani F., Le K.D., Awram P., Blake T., Nomellini J.F., Smit J. “C. crescentus Synthesizes an S-Layer-Editing Metalloprotease Possessing a Domain Sharing Sequence Similarity with Its Paracrystalline S-Layer Protein.” Journal of Bacteriology, 2002, DOI: 10.1128/JB.184.10.2709-2718.2002
10. Umelo-Njaka E., Nomellini J.F., Bingle W.H., Glasier L.G.M., Irvin R.T., Smit J. “Expression and testing of Pseudomonas aeruginosa vaccine candidate proteins prepared with the Caulobacter crescentus S-layer protein expression system.” Vaccine, 2001, DOI: 10.1016/S0264-410X(00)00362-5

