Corynebacterium renale
A Microbial Biorealm page on the genus Corynebacterium renale
.
Classification
Higher order taxa
Domain: Bacteria; Phylum: Actinobacteria; Class: Actinobacteria; Subclass: Actinobacteridae; Order: Actinomycetales; Suborder: Corynebacterineae; Family: Corynebacteriaceae; Genus: Corynebacterium [Others may be used. Use NCBI link to find]
Species
|
NCBI: Taxonomy |
Corynebacterium renale
Description and significance
The Corynebacterium genus is comprised of sixty six species (5). This large group contains human and animal, as well as, plant pathogens, and lives in a variety of habitats. One important human pathogen in the genus is C. diphtheriae which causes a fatal upper respiratory tract infection (12).
C. renale is only one species of the genus not well known or studied. The C. renale group was formerly known as immunological types I, II, and III (11). Due to the lack of genetic and chemotaxonomic identification methods, distinguishing the different species from the group remained impossible. Rather phenotypic identification of the organisms was the normal identification process used before. Only in the late 1970s, due to new methods such as genetic analyses (16S rRNA & rDNA sequencing, nucleic acid hybridization, etc), chemotaxonomic methods (detection of mycolic acids, peptidoglycan analyis, cellular fatty acid studies, menaquinone characterization, etc) and phenotypic techniques (identification systems, organic compound assimilation tests, etc) was the group distinguished as three different species (12). The organisms were then renamed C. renale (Type I), C. pilosum (Type II), and C. cystitidis (Type III) (11).
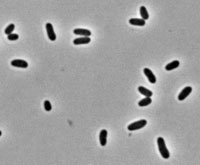
C. renale is a rather large Gram positive bacilli (0.5 by 1.3 to 2.6 μm) that is shaped as a short stumpy rod, thicker at one end then the other (11). Not surprisingly its name when defined means “club bacterium” and thus, implies this microbe’s club-like shape. In Gram stained cultures these organisms tend to clump together and form “picket fence” and “Chinese letter” arrangements (12). However, these organisms can vary from a few individuals to hundreds in colonies such as biofilms (5). Yet, C. renale is opaque, ivory-colored, and dull (11). In addition, these organisms are non-motile, non-spore bearing, and non-encapsulated (11). Through the use of their pili, they attach themselves to their favored environment in the bladder, ureters, kidney, pelvis, and kidney tissue of cows, and occasionally horses and sheep (13).
Genome structure
The complete genome of C. renale has yet to be sequenced. But due to new DNA technological advances, the 16S rRNA's genes of C. renale have been sequenced. The 16S rRNA contains approximately 1400 nucleotide bases. Moreover, through the sequencing of 16S rRNA of multiple organisms, it has led to the discovery of many new species of Corynebacterium; as well as, separated Corynebacterium species thought to be one.
In addition, C. renale contains two plasmids. This organism contains a small, cryptic plasmid called pCR1. It is the smallest corynebacterial plasmid known to date (9). This plasmid has been sequenced and contains 1441 nucleotide bases. Another plasmid C. renale has is known as pCR2. This plasmid, as well, has been sequenced and contains 3181 nucleotide bases.
Cell structure and metabolism
C. renale is a Gram positive rod. Gram positive bacteria differ from gram negative bacteria in two ways. One by having a thicker and more cross-linked peptidoglycan cell wall and two, by lacking an outer membrane (2). In particular, the peptidoglycan of C. renale contains meso-diaminopimelic acid (m-DAP) as the constituent of amino acid (5). This species also contains mycolic acids, arabinose and galactose in their cell walls, as well as, distinctive cellular fatty acids and menaquiniones (12). In addition, there are proteins anchored to this organism’s cell wall in three ways: 1) by their C-terminal ends, 2) by way of charge or hydrophobic interactions, and 3) by their N-terminal region (lipoproteins) (2). Thus, the cell wall of C. renale can be considered highly complex, because it is made up of proteins with specific binding functions and enzymatic activity that have the ability to generate energy.
Not only is the cell structure of C. renale complex, but also its metabolism. This organism uses facultative and fermentative metabolism to grow, or else it becomes inert (3). This means that this organism can make ATP by aerobic respiration when oxygen is present, but it is also capable of switching to fermentation when oxygen is not present. Most facultative anaerobes, such as C. renale, are animal pathogens. Because, this organism multiples in sterile bovine urine, it becomes strongly alkaline (11). This is because of C. renale contains the enzyme urease (11). In particular, urease catalyzes the hydrolysis of urea into carbon dioxide and ammonia. Organisms that tend to produce urease tend to be gastrointestinal or urinary tract pathogens, since urease enables them to neutralize the acid present in their acidic environments. In addition, C. renale can also perform catalase production (11). Catalase is a common enzyme that catalyzes the decomposition of hydrogen peroxide to water and oxygen. Hydrogen peroxide is a harmful by-product of many normal metabolic processes and so to prevent damage it is converted into less dangerous compounds.
Ecology
Because C. renale is a facultative anaerobe, it is an animal pathogen. Particularly in one lab study, strains of C. renale died rather quickly on media (11). Therefore, it is safe to say that resistance to physical and chemical factors in natural environments is slight. Rather, this organism is highly adapted to the bovine and ovine urinary tract (1). Moreover, Yanagawa’s study showed that the pili of C. renale are used to adhere to the vulvar epithelium of cows, sheep and horses (13). Penetration and colonization of the urinary tract is aided by the ability of C. renale to stick to aging epithelial cells. However, other predisposing factors such as pregnancy or parturition are required for the disease to occur (13).
Pathology
C. renale is the most frequently isolated member in cases of pyelonephritis (3). In general terms, pyelonephritis is an ascending urinary tract infection that has reached the kidneys. C. renale predominantly affects female animals, such as cows, sheep and horses, producing a diphtheritic inflammation of the bladder, ureters, kidney, pelvis, and frequently the kidney tissue (11). This species is highly adapted to the bovine and ovine urinary tract through its complex structure and metabolism. Particularly, its pili are used to stick adhesively to the vulvar epithelial cells of the animal’s bladder (13). In addition, the pili contain an antigenic protein which causes the inflammation of the bladder tissue (11).
Transmission between animals occurs when contaminated droplets of urine are splashed on the vulvar area of a susceptible animal from an infected carrier (11). However, research has also shown that C. renale can be found in healthy animals of diseased herds without affecting them (11).
Surprisingly, cows with pyelonephritis and urethritis have a serum antibody response to these bacteria. Organisms with urinary sediments from animals with upper urinary tract involvement were coated with the antibody IgG (11). This antibody is probably derived from the plasma that the inflammatory tissue exudates (11). The host’s protective response, however, is infective and untreated animals seldom recover. Yet, C. renale is sensitive to penicillin, streptomycin, kanamycin, erythromycin, and polymycin B (11). Therefore, penicillin in large doses is the antibiotic of choice in therapy and is often effective if administered before lesions have become far advanced.
Current Research
C. renale Contains Pili That Are pH Sensitive
In this experiment, the adhesion of C. renale bacteria to bovine epithelial cells was studied with two clones of the bacteria piliated (P+) and non-piliated (P-) (10). The percentage of cells with the attached P+ bacteria was much greater than the number of cells with attached P- bacteria (10). The adhesion of P+ bacteria was found to be pH dependent and is supported by the following facts. One that the number of attached bacteria was high at a pH above 7.6 and low at a pH below 6.8 (10). And two, the adhering P+ bacteria were eluted immediately after the pH was decreased from 7.4 to 6.4 (10). In addition, the P+ bacteria adhered to the cells immediately after the pH was increased back to 7.4 (10). This study also found that the adhesion of P+ bacteria was inhibited by anti-plus antiserum, but that it was not inhibited by amino acids, sugars, and not influenced by calcium and magnesium (10). The P- bacteria hardly attached to the epithelial cells, irrespective of the pH and other factors.
Low Iron Media Inhibits Growth of C. renale
This study was initiated to see if the iron content of the C. renale cultivation’s medium had any effect on the growth of the bacteria or their ability to produce pyelonephritis in model rats. It had previously been reported that poor or no growth of several different strains of C. renale was observed in minimal medium with added iron (4). Through the study it was observed that the iron content of the media had a definite effect on the C. renale recovered from the renal tissue of the rats (4). It was shown that C. renale was unable to produce pyelonephritis when grown in medium where the iron content was below an undetermined critical level (4). When grown in a low-iron defined medium, C. renale proved to be avirulent for rats, but when cultivated in a high iron defined medium, the organism, was extremely virulent (4). Since the iron concentration was the only factor which varied in each group, it seems reasonable to assume that iron serves a role as a “virulence factor” in producing pyelonephritis (4).
C. renale Lactate Production
In this study, acid metabolic products of 23 strains of human and animal pathogenic Corynebacterium were determined by gas chromatography (8). The results showed that each species was metabolically the same, and each can be identified based on their produced acid products (8). C. renale was shown to produce lactate (8). Specifically, C. renale is the only Corynebacterium that produces alkaline coagulation and peptonization in litmus milk (8). The study showed that this organism is quite distinct from others in its process of producing lactic acid. One of the two strains tested (ATCC 19412) also produced trace amounts of acetate, propionate, pyruvate and succinate (8).
References
1. Baron, Samuel, MD, Milton R.J. Salton, and Kwang-Shin Kim. Medical Microbiology. 4th ed. Galveston: University of Texas Medical Branch, 1996. NCBI. 23 Aug 2007. http://www.ncbi.nlm.nih.gov/books/bv.fcgi?rid=mmed.section.284.
2. Fischetti, Vincent A, Richard P. Novick, Joseph J. Ferretti, Daniel A. Portnoy, & Julian I Rood, ed. Gram Positive Pathogens. 2nd ed. Washington D.C.: ASM Press, 2006.
3. Forbes, Betty A., Daniel Sahm and Alice S. Weissfeld. Bailey and Scott’s Diagnostic Microbiology. 11th ed. St. Louis: Mosby, 2002.
4. Henderson, Lyn C, Solomon Kadis and Willie L. Chapman Jr. “Influence of Iron on Corynebacterium renale-Induced Pyelonephritis in a Rat Experimental Model.” Infection and Immunity 21.2. (1978). PubMed Central. 23 Aug 2007. http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=422029&blobtype=pdf.
5. Mahon, Connie R, Donald C. Lehman, & George Manuselis. Diagnostic Microbiology. 3rd ed. St. Louis: Saunders Elsevier, 2007.
6. Olson, Merle E, Howard Ceri, Douglas W. Morck, Andre G. Buret, and Ronald R. Read. “Biofilm bacteria: formation and comparative susceptibility to antibiotics.” Canadian Journal of Veterinary Research 66.2. (2002). PubMed Central. 23 Aug 2007. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=226988.
7. Reddy, Adinarayana and Marina Kao. “Value of Acid Metabolic Products in Identification of Certain Corynebacteria.” Journal of Clinical Microbiology 7.5: 428-233 (1978). PubMed Central. 23 Aug 2007. http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=275008&blobtype=pdf.
8. Salkinoja-Salonen, M. S, R. Vuorio, M. A. Anderson, P. Kampfer, M. C. Andersson, T. Honkanen-Buzalski and A. C. Scoging. “Toxigenic Strains of Bacillus licheniformis Related to Food Poisoning.” Applied and Environmental Microbiology 65.10. (1999): 4637–4645. PubMed Central. 23 Aug 2007. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=91618.
9.Srivastava, P, Nath N, and Deb JK. “Characterization of Broad Host Range Cryptic Plasmid pCR1 from Cornyebacterium renale.” Plasmid 56.1: 24-34. (2006). Pub Med. 23 Aug 2007. http://www.ncbi.nlm.nih.gov/sites/entrez?Db=pubmed&Cmd=ShowDetailView&TermToSearch=16545871&ordinalpos=4&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum.
10. Takai, S, Ryo Yanagawa and Yukitoshi Kitamura. “pH-dependent Adhesion of Piliated Corynebacterium renale to Bovine Bladder Epithelial Cells.” Infection and Immunity 28.3: 669-674 (1980). PubMed Central. 23 Aug 2007. http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=551002&blobtype=pdf.
11. Timoney, John F, James H. Gillespe, Frederic W. Scott and Jeffrey E. Barlough. Hagan and Bruner's Microbiology and Infectious Diseases of Domestic Animals. Ithaca: Cornell University Publishing, 1988.
12. Winn Jr, Washington, Stephen Allen, William Janda, Elmer Koneman, Gary Procop, Paul Schreckenberger, and Gail Woods. Koneman’s Color Atlas & Textbook of Diagnostic Microbiology. 6th ed. Philadelphia: Lippincott Williams & Wilkins, 2006.
13. Yanagawa, Ryo and Koichi Otsuki. “Some Properties of the Pili of Corynebacterium renale.” Journal of Bacteriology 101.3. (1970): 1063-1069. PubMed Central. 23 Aug 2007. http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=250427&blobtype=pdf.
Edited by a student of Rachel Larsen
Edited by KLB