Saccharomyces
A Microbial Biorealm page on the genus Saccharomyces
Classification
Higher order taxa
Eukaryota; Fungi/Metazoa group; Fungi; Ascomycota; Saccharomycotina; Saccharomycetes; Saccharomycetales; Saccharomycetaceae
Species
Saccharomyces cerevisiae
Saccharomyces boulardi
Saccharomyces uvarum
Description and Significance
The genus Saccharomyces are unicellular fungi, or "yeasts." The name Saccharomyces means "sugar fungus." S. cerevesiae is also known as brewer's yeast for brewing beer.
Many signalling pathways of universal importance in eukaryotes were first discovered in Saccharomyces. Retrograde signaling, which consists of signaling pathways leading from the mitochondria to the nucleus, was first discovered in Saccharomyces cerevisiae. To protect itself from acetic acid-mediated programmed cell death, Saccharomyces cerevisiae utlizes acid stress adaptation. The response to a programmed cell death by acetic acid is mediate through ROS-dependent apoptotic pathways. A study by Giannattasio et. al. (2005) gave some evidence that H2O2 detoxification may play a role in preventing yeast cell death in response to acetic acid.
Genome Structure
Chromosomes of Saccharomyces contain a single linear double-stranded DNA with few repreated sequences. It is believed that these repetitions are caused by the encoding of ribosomal RNA. Less than 5% of sequences have introns. The Saccharomyces cerevisiaegenome was completed in 1996. This species has 6,000 genes.
Because of its genetic structure, Saccharomyces cerevisiae is a useful research organism. For example, scientists at the Woolford Laboratory at Carnegie Mellon University have used it to study pathways of ribosome assembly. This in turn has led to a better understanding of the genetic structure not just of this organism, but to certain genetic processes in general. Like other eukaryotes, the 40S ribonucleoprotien contains one 18S rRNA and 32 ribosomal protiens. These and other rRNA structures come from a single 35S transcript. This transcript is synthesizes by polymerase I, while pre-RNA is transcribed by polymerase III. The pre-RNA is later packaged in a 90S RNP. These processes are mediated by endoribionucleases and exoribionucleases. After assembly, 66S particles are released into the nucleoplasm, mature, and are exported to the cytoplasm. However, Woolford Laboratory researchers have discovered that there are still other steps before ribosomal subunits are able to facilitate protein synthesis. They have also noted non-ribosomal molecules, which are necessary for processes such as rearrangement of rRNA structure as well as RNA cleavage and processing.
Other research has been done on the genetic structure of Saccharomyces cerevisiae. For example, Harry Alices-Villanueva (1997) studied the TRP1 RI cirlce plasmid, found on chromosome IV of this species. The TRP1 gene in the plasmid codes for an ezyme required in the synthesis of tryptophan. This plasmid contains the Autonomously Replicating Sequence (ARS). The ARS allows the plasmid to replicate independently of chromosomal DNA. An important feature of the TRP1 RI circle is that it has a high copy number. Alices-Villanueva's research tested the theories about why this high copy number occurs. Specifically, he looked at a hypothesis which states that TRP1 gene includes a weak minor promoter instead of a strong major one. Alices-Villanueva created two versions of this gene, one with only a strong promoter and one with both a strong and a weak promoter. Analysis showed rates of expression higher for the gene with both promoters, giving evidence to the proposed hypothesis.
Saccharomyces cerevisiae contains an acidic cytoplasmic protien named Gir2. Alves and Castilho (2005) have suggested that this protein lacks extensive seconday structure. In a recent study, they reported that Gir2 is sensitive to proteolysis. Saccharomyces cerevisiae also contains the gene JEN1. S. cerevisiae rho(0) strains do not have the entire mitochondrial genome. Certain nuclear genes in Saccharomyces cerevisiae respond to mitochondria in the Butow retrograde regulation process. This is activated in cells that lack mitochondrial involvement of certain genes. Recently, Kelberg et. al. (2005) discovered a new gene in Saccharomyces cerevisiae called HIM1. They mapped this gene on the right arm of chromosome IV. When mutations in HIM1 occur, there was an increase both in spontaneous mutation rate and in overall frequencies of mutations.
Cell Structure and Metabolism
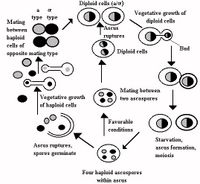
Most Saccharomyces species are heterothallic, but a few are homothallic. If sexual reproduction occurs, vegetative cells act as asci. Four ascospores are formed during meiosis. Once released, these ascospores germinate, producing haploid strains. To return to the diploid state, mating between haploid cells must occur. Haploid and diploid phases are morphologically similar, but diploid cells are larger. In asexual reproduction, bud grows to reach the size of the mother cell while nuclear division occurs. After a nucleus is passed to the daughter cells, separation occurs.
Saccharomyces cells are typically oblong spheroids with a prominent central vaculoe and a small nucleus. Glucan is a major component of cell walls, as well as mannoprotiens. Ascospores contain chitosan and dityrosine. Cells grow by budding.
Saccharomyces have an active glucose transport system. It metabolizes glucose through the glycolytic pathway. This system is effective at low glucose concentrations, but if concentrations are high, the system will be repressed. If repression occurs, glucose enters the cell via a constitutive falilitated diffusion system. High glucose levels may also suppress respiration in favor of fermentation, even if oxygen is available. This is known as catabolite repression or the Crabtree effect. Saccharomyces may be inducible.
Saccharomyces are heterotrophes, obtaining energy from glucose. They utlize both respiratory and fermentative metabolism. Roughly 98% of glucose is metabolized during fermentation, while 2% of it is made into cell materials. Anaerobic metabolism, however, yields more energy, so 10% of the glucose can be converted to cell material. This pheonomena is known as the Pasteur effect.
Ecology
Saccharomyces cerevisiae's natural habitat is on the surface of fruit, but it is best known for its role in the baking and brewing industries. This species is considered an ale yeast, also known as a top yeast. This means that during fermentation the yeast mixes with gas and accumulates at the top of the vat. Another strain of this species, Saccharomyces carlsbergensis, is also used in brewing. It is considered a lager yeast, or a bottom yeast. This yeast sedmients instead of rising to the top of the brew. Yeast cells used in brewing produce compounds for flavor and taste such as ethyl esters and dimethyl sulphide. It is easy to remove yeast cells after fermentation, because they flocculate and clump as the process ends. These cells can be recylced, although eventually must be discarded to avoid contamination. The used yeasts can be recycled even further by using them in animal feed or as a source of vitamins.
S. cerevisiaehas medical applications. It is sometimes used as a probiotic in humans. However, research has shown that use of S. cerevisiae probiotics may actually be harmful. Munoz et. al. (2005) documented an emerging infection called S. cervisiae fungemia. Their research indicated that the only apparent risk factor for this disease is having been treated with a probiotic made from Saccharomyces boulardii. This probiotic is used in Europe to treat Clostridium difficile-associated diarrhea. Based on their findings, Munoz et. al. concluded that the use of these probiotics should be reconsidered.