Streptococcus
A Microbial Biorealm page on the genus Streptococcus
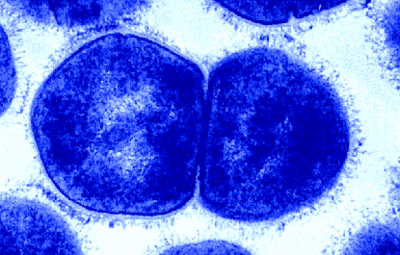
Numbered ticks are 11 µM apart.
Gram-stained.
Photograph by Bob Blaylock.
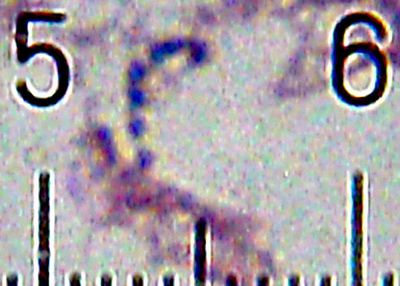
Numbered ticks are 11 µM apart.
Gram-stained.
Photograph by Bob Blaylock.

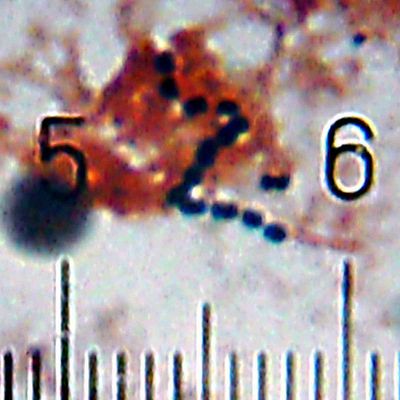
Numbered ticks are 11 µM apart.
Gram-stained.
Photograph by Bob Blaylock.
Classification
Higher order taxa:
Bacteria; Firmicutes; Bacilli; Lactobacillales; Streptococcaceae; Streptococcus
Species:
Streptococcus acidominimus, S. agalactiae, S. alactolyticus, S. anginosus, S. anthracis, S. australis, S. bovis, S. canis, S. castoreus, S. constellatus, S. criae, S. criceti, S. cristatus, S. devriesei, S. didelphis, S. downei, S. dysgalactiae group, S. entericus, S. equinus, S. ferus, S. gallinaceus, S. gallolyticus, S. genomosp. C1, S. genomosp. C2, S. genomosp. C3, S. genomosp. C4, S. genomosp. C5, S. genomosp. C6, S. genomosp. C7, S. genomosp. C8, S. gordonii, S. halichoeri, S. hyointestinalis, S. hyovaginalis, S. infantarius, S. infantis, S. iniae, S. intermedius, S. luteciae, S. lutetiensis, S. macacae, S. macedonicus, S. marimammalium, S. massiliensis, S. milleri, S. minor, S. mitis, S. mutans, S. oligofermentans, S. oralis, S. orisratti, S. ovis, S. parasanguinis, S. parauberis, S. pasteuri, S. pasteurianus, S. peroris, S. phocae, S. pluranimalium, S. pneumoniae, S. porcinus, S. pseudopneumoniae, S. pyogenes, S. ratti, S. salivarius, S. sanguinis, S. sciuri, S. sinensis, S. sobrinus, S. suis, S. thermophilus, S. thoraltensis, S. uberis, S. urinalis, S. vestibularis, S. viridans, S. sp
Description and Significance
Streptococci are a diverse genus, infecting a barrage of different animals, including humans, with diseases ranging from strep throat to necrotizing fasciitis. They have come to public attention recently as antibiotic-resistant strains have started appearing and causing epidemics. In an effort to battle the evolution of these clever pathogens, researchers have sequenced the genomes of 9 different strains in 4 different species of Streptococcus.
Genome Structure
The gene sequence for the circular chromosomes of twelves pecies of streptococci have been sequenced: Streptococcus pneumoniae, Streptococcus agalactiae 2603 V/R , Streptococcus mutans, Streptococcus thermophilus CNRZ1066, Streptococcus thermophilus LMG 18311, and Streptococcus pyogenes. The genome of Streptococcus pyogenes is 1,852,442 base pairs long, containing 1,752 predicted protein-coding genes. Researchers have identified more than 40 virulence-associated genes on the S. pyogenes genome, which fits in with its ability to cause a variety of human diseases. Researchers also found numerous genes that encode proteins that aid in "molecular mimicry," when the bacterium disguises itself by posing as a cell that is a part of the host.
The genome of S. pneumoniae is 2,038,615 base pairs long with 2,043 predicted protein-coding regions. S. pneumoniae has a high capacity for DNA uptake, and consequently its genome contains many genes that are most likely derived from other bacteria. More than 53% of S. pneumoniae genes have extremely similar counterparts on the Bacillus subtilius genome. Both are very prototypic Gram-positive bacteria.
The genome of S. agalactiae contains 2,160,167 base pairs long with 2,175 predicted protein-coding regions. The genome of S. agalactiae is very similar to other Streptococcus species such as S. pyogenes and S. pneumoniae, but differs in several metabolic pathways and membrane transport systems. These differences are attributed to adaptations to host environment.
The genome of S. mutans contains 2,030,936 base pairs with 1,963 predicted protein-coding regions. S. mutans is the leading cause of tooth decay worldwide. S. mutans is capable of metabolizing a wider variety of carbohydrates than any other Gram-positive organism yet sequenced. S. mutans also has an extensive transport system (coding takes up around 15% of the genome) that it uses for the uptake of sugars from the environment. Clearly S. mutans is highly dependent upon its host for resources.
Cell Structure and Metabolism
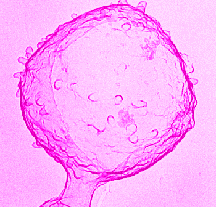
Streptococci are nonmotile, Gram-positive, nonsporeforming bacteria, that live in pairs or chains of varying length. They are characteristically round or ovoid in shape. Most Streptococci are facultative anaerobes, although some are obligate anaerobes. They usually require a complex culture medium in order to grow. Many streptococci imitate aspects of their host in order to escape detection. The capsule of Streptococcus pyogenes is chemically similar to that of it's host's connective tissue, and therefore, is nonantigenic, and it's cytoplasmic membrane has antigens similar to human cardiac skeletal and smooth muscle.
Ecology
Streptococci are a part of normal animal flora. Although some can cause diseases. The progression from latency to virulence is not well-understood, but the sequencing of the Streptococcus genomes is aiding researchers in understanding better the mechanisms of streptococci.
Pathology
Infections by Streptococci are separated into several categories, depending on the composition of their cell walls. Groups A and B are the most common and devastating human pathogens. Group A Streptococcus bacteria causes disease ranging from streptococcal sore throat (strep throat) to necrotizing fasciitis (flesh-eating disease). They can also cause scarlet fever, rheumatic fever, postpartum fever, and streptococcal toxic shock syndrome. S. pyogenes can be counted among their numbers as one of the major pathogens in group A streptococci. Necrotizing fasciitis is one of the most deadly strep infections, due to its rapid progression. It is an infection caused by a deadly strain of group A strep that attacks the deep layers of tissue (fascia). The strain is normally not so aggressive, and it is thought that its sudden virulence is triggered by lateral gene transfer by a bacteriophage.
Streptococcal toxic shock syndrome is another serious strep infection that progresses very rapidly. It causes a dangerous drop in blood pressure, damage to the kidneys, liver, and lungs, and eventually shock. Due to its rapid progression, the damage is usually done before the disease can even be diagnosed, let alone treated.
Group B streptococci cause life-threatening diseases in newborns, pregnant women, the elderly, and adults with compromised immune systems. Group B strep infections are different from other strep infections, in that the individual can be colonized by the bacteria before any symptoms are obvious. This means that people can carry the bacteria in their bodies but are not infected, and do not show any symptoms. Group B strep can be carried in the gastrointestinal tract, genital tract, or urinary tract, and only become dangerous when they invade the bloodstream.
Among group B infections is pneumonia. Pneumonia can be caused by a barrage of different things including viruses and fungi, but is most commonly caused by Streptococcus pneumoniae (mentioned above in the genome section) also called pneumococcus, which is the only type of pneumonia for which there is a vaccine. S. pneumoniae are often present in healthy throats and only develop into a serious infection when the host's defenses are depleted due to such factors as old age, illness (i.e. AIDS), or malnutrition. Like necrotizing fasciitis and toxic shock syndrome, bacterial pneumonia progresses vary rapidly with a sudden onset of high fever. More seriously, the infection can become invasive and manifest itself as meningitis (an infection of the cerebrospinal fluid).
Medicine
Streptococci have been the focus of a lot of medical research because of newly emerging, antibiotic-resistant strains. Research into new antibiotics to treat the diseases and new vaccines to prevent them has escalated in recent years. And information on streptococci has increased with the sequencing of the genome of four different species of streptococci. Streptococci have been shown to be resistant to erythromycin and penicillin, but they have also been shown to resist macrolide antibiotics. But low and prolonged exposure to the macrolide antibiotics seem to have allowed the streptococci to evolve a resistance to themselves, but this also made them resistant to penicillin and erythromycin. In a study the centers that had streptococci with a high macrolide resistance also had a penicillin resistance.
Streptococcus in the news
Streptococcus suis, a species of Streptococci found in pigs, has recently caused a rash of human infections in China. Human-to-human contact does not seem to be the bacteria's mode of transport, rather, Streptococcus suis is contracted by contact with or the consumption of infected pigs. As of now the death toll from this illness is 37, an unusually high mortality rate causing some scientists to suspect that swine flu may not be the disease. The known vaccines have not been produced in China for some time because of a lack of demand, but currently vaccine production is underway in light of the disease outbreak. (Information from Reuters Alert Network)
References
ASTDHPPHE: Group A and B Streptococcus
Bacteriology at UW-Madison: Streptococcus pyogenes
Center for Disease Control and Prevention: Streptococcus pneumoniae
eMedicine: Necrotizing fasciitis[1]
Science Education: Necrotizing fasciitis: "the flesh-eating bacteria"
Tettelin,Herve et al. 2002. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. National Academy of Science, Vol. 99: 12391-12396.
