The Relationship Between Ebola Virus and Host Factors
By: Amanda He (currently being edited)
Introduction to Ebolavirus

Image source: http://viralzone.expasy.org/all_by_species/207.html
Ebolaviruses (EBOV) are enveloped viruses that belong to the Filoviridae family. EBOV has received a significant amount of international attention because it causes severe hemorrhagic fevers in humans with a fatality rate of approximately 60% [1]. A minority of cases with infection results in flu-like symptoms with the addition of mild blood coagulopathy, blood loss, and increase in white blood cells, but these patients have a full recovery. The majority of cases develop severe illness with excessive hemorrhaging and blood clotting, which results in shock and death [1]. However, it is important to note that a major factor of the fatality rate of Ebolavirus disease is the access to proper healthcare. EBOV is extremely infectious and can be transmitted through exposure to bodily fluids such as feces, saliva, urine, vomit, and semen from an infected individual and can enter the body through broken skin or unprotected mucous membrane. For a healthcare worker who may be interacting with a number of different patients, it is important to wear appropriate protective equipment and practice appropriate infection control and sterilization measures [2]. Potentially infected individuals must be isolated from other patients to prevent further outbreaks. Although an individual may be infected, the symptoms are not automatic as the virus has an incubation period ranging from 2 to 21 days [3]. The average incubation period is 7-10 days. Through isolation of a potentially infected individual, it can prevent the spread to others.
Several studies have characterized EBOV infection to disable the immune system and the vascular system [3]. The disabling of the vascular system is what leads to the symptoms such as hemorrhage, hypotension, and blood pressure drop [3]. Autopsy of EBOV infected patients found the viruses located primarily in the endothelial cells, mononuclear phagocytic cells, and several fibroblasts and hepatic sinusoids [3]. As a result, it is likely that the initial infection of EBOV targets macrophages and monocytes [4]. The infection of macrophages and monocytes leads to increased synthesis of tumor necrosis factor-α (TNF-α), which induces fever and lymphoid cell apoptosis [3]. Additionally, infection of macrophages and monocytes can induce the release of various pro-inflammatory proteins.
As of 2014, there are no approved therapeutic strategies to treat infection. As a result, there has been a significant emphasis on studying the host factors EBOV recruits [5]. Host factors are traits found in an individual that could have an effect on their susceptibility to disease. In recent studies, certain host factors such as cathepsin B, heat shock 70 kDa protein 5, and STAT1 have been identified to play a role in EBOV infection. Further investigation of host factors can help grasp a better understanding about EBOV and potentially lead to a treatment.
Ebola Genome and Replication Cycle
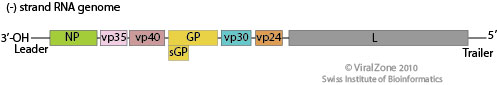
Image source: http://viralzone.expasy.org/all_by_species/207.html
EBOV has a filamentous shaped virus with a lipid envelope, lipid bilayer coat, and genome. The lipid bilayer coats serves as a mechanism to protect the genome and assists with entry into the host cell [6]. The genome of EBOV consists of single stranded, nonsegmented, negative-sense RNA virus with approximately 19,000 bases [7]. The EBOV genome encodes for seven proteins: glycoprotein (GP), matrix protein viral protein 40 (VP40), nucleoprotein (NP), viral protein 24 (VP24), viral protein 30 (VP30), viral protein 35 (VP35), and large protein, an enzyme subunit (L) [1].
Each of these proteins play a significant role in EBOV infection. GP is found on the viral envelope and plays a role in viral attachment and entry into the host cell [7]. VP40 is found in the viral lipid coat and plays a role in virus structure and stability [1]. Researchers found that with the expression of only one EBOV protein, VP40, the virus is still able to form virus like particles in human cells. All the remaining proteins are used to compose the nucleocapsid. The nucleocapsid is the capsid surrounding the RNA, which plays an important role in viral transcription and replication.
Host Factors: HOPS and NPC1
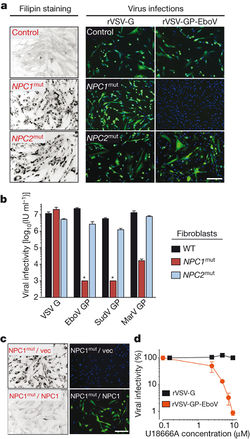
A) Fluorescence microscopy viewing of skin fibroblasts with NPC1 and NPC2 mutations and the susceptibility to EBOV infection.
B) Quantification of infectivity from fluorescence microscopy images.
C) NPC1 patient fibroblasts challenged with rVSV-GP-EBOV.
D) Percentage of Vero cells infected with increasing concentrations of U18666A.
Image source: http://www.nature.com/nature/journal/v477/n7364/full/nature10348.html [8]
In a genome-wide haploid genetic screen to identify host factors required for EBOV entry, the most mutations found were in homotypic fusion and vacuole protein-sorting (HOPS) multisubunit tethering complex and Niemann-Pick C1 (NPC1). HOPS is involved in fusion of endosomes to lysosomes and plays a role in endosome maturation, while NPC1 is an endo/lysosomal cholesterol transporter protein that can affect endosome/lysosome fusion and fussion and calcium homeostasis [8].
Carette et al took skin fibroblast samples from healthy individuals and tested the susceptibility to Filviridae infection, which typically utilizes the GP protein to enter the host cell [8]. Since both NPC1 and NPC2 are cholesterol transporter proteins, both were used and mutated to not fully express. Although both NPC1 and NPC2 had identical cholesterol levels as shown through filipin staining, only the NPC1 mutant was less susceptible to infection to EBOV [Fig 3A,B]. NPC2 had similar infection percentages as the wild-type [Fig 3B]. The researchers wanted to confirm that the NPC1 is required for EBOV GP-mediated infection, so they used U18666A, a small molecule that causes NPC1 deficiency by targeting NPC1 [8]. When Vero and HAP1 cells were exposed to increasing concentrations of U18666A, it was able to inhibit viral infection of EBOV-GP, but not the rabies virus [Fig 3D]. From this result, there is a likely chance that NPC1 may have an interaction with EBOV GP-protein that allows it to infect the host cell.
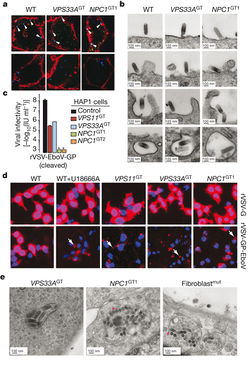
A) Identification of viral particle interaction and infection in HOPS and NPC1 deficient cells. EBOV = blue.
B) Electron microscopy visualization of EBOV entrance into cells.
C) Quantification of viral infectivity in various mutated strains.
D) Fluorescence microscopic images of the effect of U18666A or mutations in HAP1 cells on EBOV infectivity.
E) Electron micrographs of EBOV infected mutant strains and NPC1 deficient fibroblasts.
Image source: http://www.nature.com/nature/journal/v477/n7364/full/nature10348.html [8]
Once it was confirmed that NPC1 and HOPS complex were needed for virus infection, a series of experiments ranging from virus binding to VSV M protein-release assay to electron microscopy were run to identify how virus infection would be inhibited in cells deficient for HOPS complex and NPC1. The HAP1 cells were mutated to generate, VPS33AGT and VPS11GT – the HOPS knockout strains, and NPC1GT – the NPC1 knockout strain. Quantifying the amount of virions that were able to bypass the infection block that the knockout strains established, they found that both knockout strains had a significantly lower total amount of cellular infection from EBOV-GP in comparison to the control, HAP1 cell [Fig 4C]. However, NPC1 knockout strains had the lowest amount of infection as they had roughly 62% less infection than the HAP1 cells, while the HOPS knockout strains had roughly 37% less [Fig 4C].
In Fig 4D, viral escape into the cytoplasm was measured. The presence of red surrounding the blue of the viral particles indicates membrane fusion [8]. In the wild-type cells that were not treated anything, there is a significantly high amount of red throughout when exposed to EBOV, which means that there is more viruses escaping the cell [Fig 4D]. However, the wild-type cells that are treated with U18666A have reduced the number of viruses escaping. The U18666A treated cells had identical results to the HOPS knockout strain and the NPC1 knockout strain. Overall, the results consistently confirm that HOPS and NPC1 are somehow used by EBOV to enter and exit host cells. Through knocking out the complexes or using inhibitory molecules, there was a prevalent decrease in the number of infected cells. It could be of potentially interest to look into the NPC1 gene for EBOV therapeutics as it was roughly twice as effective as HOPS.
Host Factor: HSPA5
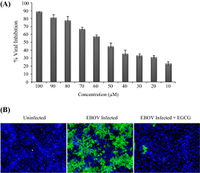
A) Percentage of EBOV infection inhibition relative to EGCG concentration.
B) Visualization of immunofluorescence detection of EBOV infection.
Image source: http://www.sciencedirect.com/science/article/pii/S0166354214002034 [5]
An endoplasmic reticulum (ER) chaperone called heat shock 70 kDa protein 5 (HSPA5) has been identified as an EBOV-associated host factor [9]. HSPA5 is a highly conserved ER protein that assists with protein folding and assembly. It has additional roles in regulation of ER stress responses. However, various viruses are able to utilize HSPA5 for their own purposes such as assistance in development of mature viral envelope proteins and viral entry [5]. It is likely that HSPA5 may be utilized by EBOV assist in infection.
In Reid, et al, 2014, the laboratory members investigated how inhibition of HSPA5 affects EBOV infection. In order to verify the effects of HSPA5 on EBOV infection, the researchers detected the percentage of cells infected with EBOV when HSPA5 activity was inhibited at the ATP-binding site. The inhibitor that was used is called epigallocatechin gallate (EGCG), an antioxidant that prevents damaging effects of free radicals. EGCG is also a molecule that successfully binds to the ATP-binding site of HSPA5, hindering its functions. In the experiment, EGCG was introduced into HeLa cells at varying concentrations and incubated for two hours prior to infection with EBOV. Two days after EBOV infection, the cells were fixed and stained for with Hoechst 33342 [Fig 4]. Using immunofluroscence imaging, they identified the identified the significant decrease in viral infection. All cell nuclei were stained blue, but infected cell nuclei would fluoresce a green color [Fig 5B]. Quantifying the amount of cells that fluoresced green in proportion to the blue uninfected cells, there was a direct correlation between the levels of EGCG to the percentage of EBOV inhibition. There was a consistent pattern of increased EBOV inhibition with increased concentration of EGCG [Fig 5A]. The results showed promise that HSPA5 and interaction with its ATP-binding site play a role in EBOV replication.
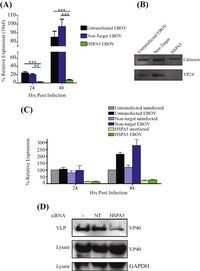
A) Expression of EBOV mRNA with non-target and HSPA5 siRNAs 24 and 48 hours post-infection.
B) Western blot of amount of viral protein 24 (VP24) produced 48 hours post-infection.
C) Expression of HSPA5 mRNA in cells infected and uninfected by EBOV 24 and 48 hours post-infection.
D) WEstern blot of amount of viral protein 40 (VP40) produced 48 hours post-infection.
[5
As there are known viruses that utilizes HSPA5 for viral protein maturation and viral entry, further experiments were ran to identify the protein production when HSPA5 is inhibited. 293T cells were treated with HSPA5 target siRNA and then infected with EBOV for 24 or 48 hours. RNA was isolated and quantified for presence of viral and HSPA5 mRNA [Fig 6A,C]. Only the 293T cells that had HSPA5 knocked out had significantly lower relative expression of EBOV mRNA, while the untransfected control and non-target siRNA control both had similar high levels of expressed EBOV mRNA [Fig 6A]. When used in a Western blot, they were able to identify that without the expression of HSPA5, there was VP24 formation and reduced formation of VP40 [Fig 6B,D]. The lack of HSPA5 preventing the formation of VP24, a protein involved in suppressing type I interferon production, and VP40, a protein involved in viral budding, shows that it is utilized by EBOV in a similar fashion to other viruses [5].
A takeaway from the Reid, et al study is the possibility of targeting HSPA5 to inhibit viral infection of EBOV instead of the EBOV genome. In both in vitro and in vivo experiments targeting HSPA5 resulted in significant reduction in virus replication and higher survival of mice against the lethal dosage of EBOV. By targeting a host factor instead of the viral genome, there would be greater difficulty for the virus to make escape mutants [5].
Host Factor: KPNA5
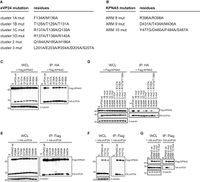
List of mutations in A) VP24 residues and B) KPNA5 ARMS. Co-immunoprecipitation of C) VP24 single residue and D) multiple-residue VP24 mutants.
Image source: http://www.plospathogens.org/article/info%3Adoi%2F10.1371%2Fjournal.ppat.1002550 [10]
Interferons are proteins created and released in host cells in response to pathogens. Interferons are able to generate innate and adaptive immune responses to viral infections by signaling to signal transducer and activator of transcription (STAT) family transcription factors [Xu 2014]. Signal transducer and activator of transcription 1-α/β (STAT1) is a member of the family that has been found to interact with the EBOV viral protein 24 (VP24) [10, 12]. Viral protein 24 (VP24) is one of the eight proteins that EBOV encodes in its genome. It has a number of different roles ranging from virus replication and assembly to binding to assisting in immune suppression [10]. Through X-ray crystallography, VP24 has been shown to bind directly to STAT1. In Xu, et al, 2014, the researchers were able to identify the binding site on STAT1 that VP24 binds to.
VP24 interacts with STAT1, phosphorylated at tyrosine 701 (PY-STAT1) because STAT proteins often undergo tyrosine-phosphorylation to form homodimers and heterodimers [12]. The dimer formations can interact with interferon / expression [11]. The structure of PY-STAT1 can be recognized by karyopherin alpha (KPNA) nuclear transport factors. The binding of KPNA factors mediate nuclear transport without impacting nucleocytoplasmic trafficking processes [12]. Many viruses inhibit STAT1 activation in order to target interferon signaling, so it is likely that EBOV has a specific target on STAT1 as well [11].
In order to identify how VP24 promotes immune suppression, Xu, et al, 2014 characterized the interaction between VP24 and KPNA5, a transport factor that binds specifically to PY-STAT1 with nuclear translocation. From X-ray crystallography, three clusters of VP24 were found to have contact with KPNA5 [12]. There were extensive hydrogen bonding and hydrophobic interactions between VP24 and KPNA5 ARM 8, 9, and 10 [12]. The complex indicated that there were a large number of VP24 residues at the binding interface that interact with KPNA5 interactors. With the three identified clusters, clusters 1 and 3 have been conserved amongst different EBOV and Marburg viruses [12]. The conserved nature of these clusters may indicate that they serve as a specificity determinant of VP24. In order to determine the selectivity, the researchers introduced mutations of the VP24 interaction residues [Fig 7A] and the KPNA5 ARMs [Fig 7B]. All point mutations in VP24 residues, with the exception of arginine 137 to alanine (R137A) showed a < 20% loss of KPNA5 binding [Fig 7C]. In KPNA5 mutations, only four mutants (R398A, Y477A, Y477G, and F484A) had an effect on the binding affinity to KPNA5 [Fig 7E, F]. From these results, they were able to confirm that VP24 and KPNA5 share a unique binding interface. In later experiments, they discovered that PY-STAT1 shares the exact same binding spot with VP24 on KPNA5 [12].
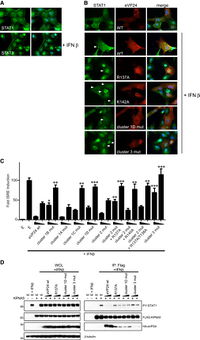
A) Identification of STAT1 nuclear accumulation with and without interferon-beta.
B) Ectopic expression of VP24 WT or mutants relative to PY-STAT1 translocation to nucleus.
C) The rate at which VP24 was able to inhibit induction of ISG54 promoter. Error bars = SEM.
D) Competition between VP24 and PY-STAT1 for KPNA5 binding at 2 ug and 4 ug.
Image source: http://www.plospathogens.org/article/info%3Adoi%2F10.1371%2Fjournal.ppat.1002550 [10]
Next, they investigated how VP24 binding to KPNA5 affected STAT1-mediated nuclear transport and signaling. Fluorescence imaging was utilized to confirm the effect of interferon-β on STAT1 as its presence lead to nuclear accumulation of STAT1 [Fig 8A]. Then, they competed the VP24 mutants with reduced KPNA5 binding activity against PY-STAT1 to identify if there was an effect on PY-STAT1 nuclear trafficking [Fig 8B]. When interferon-β was expressed, there was a significant reduction in PY-STAT1 inhibition to nuclear translocation, indicating that the weaker bond between VP24 and KPNA5 had weaker inhibition of PY-STAT1 movement. In Fig 8D, they tested a direct competition model in order to identify VP24 wild-type and its ability to competitively bind to KPNA5, inhibiting PY-STAT1 and KPNA5 binding. The results indicated that VP24 and its mutants seem to have an equal probability to interact with KPNA5 as PY-STAT1 does [Fig 8D].
From the results, it was confirmed that VP24’s binding to KPNA5 is a competitive inhibitor action. By binding to KPNA5, there would be a reduction in PY-STAT1 nuclear localization, which hinders the STAT1’s ability to establish cell-intrinsic antiviral signaling [12]. As a direct result, EBOV is able to facilitate viral replication. Further investigation of the binding between STAT1 and VP24 must be conducted in order to establish a potential drug that would prevent VP24 from binding to KPNA5. Once VP24 is no longer binding,
Host Factor: TOP1
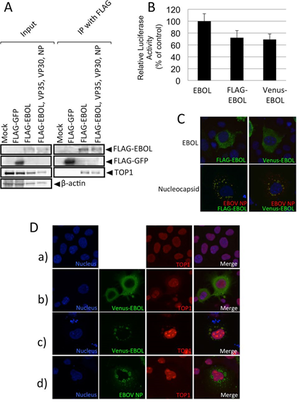
A) Cells expressing FLAG-EBOL immunoprecipitated and Western blotted for FLAG and TOP1.
B/C) Verification of similar polymerase and intracellular localization between FLAG-EBOL and Venus-EBOL.
D) Intracellular localization of TOP1 in EBOV protein-expressing cells. TOP1 = red. Nuclei = blue.
Image source: http://jvi.asm.org/content/87/16/8862.full [13]
In EBOV, L-protein serves as a viral RNA-dependent RNA polymerase [13]. In a study to identify what proteins interacted with the L-protein (EBOL), Takahashi, et al, 2013 ran several minigenome and coimmunopreicipitation assays on EBOL plasmid, tagged with the green fluorescent protein at the N-terminal, FLAG (FLAG-EBOL). They were able to identify 65 different cellular proteins that interacted with the EBOL. DNA topoisomerase 1 (TOP1) was one of the more interesting proteins discovered. DNA topoisomerase 1 (TOP1) is a nuclear protein interacts with both DNA and RNA. In double-stranded DNA, TOP1 binds to the helical structure and assists in unwinding the DNA [13]. In RNA, it recognizes stem-loop RNA structures [13]. Since EBOV genome is RNA, it is possible that TOP1 played a role in the transcription and replication of EBOVA RNA genome.
In order to verify the interaction between EBOL and TOP1, Takahashi, et al, 2013 performed an immunoprecipitation assay and Western blotting [Fig 9A]. TOP1 was successfully coimmunoprecipitated with the tagged EBOV genome, indicating that the two do interact. Next, they wanted to identify the location at which TOP1 and L-protein interacted. In order to visualize it, the EBOL plasmid was tagged with another green fluorescent protein, Venus (Venus-EBOL), which showed similar characteristics of polymerase activity to FLAG-EBOL [Fig 9B, C]. Nuclei were stained with Hoechst stain (blue) and TOP1 was identified by red fluorescence of anti-TOP1 antibody. In the mock-treated cells, TOP1 was detected only in the nucleus as shown by the purple merged image [Fig 9D-a]. Cells transfected with Venus-EBOL had TOP1 located in both the nucleus and the cytoplasm [Fig 9D-b]. Cells transfected with Venus-EBOL and VP30, VP35, and NP had TOP1 detection in inclusion bodies and the cytoplasm [Fig 9D-c]. Meanwhile in cells transfected with only VP30, VP35, and NP, there was only TOP1 found in the nucleus [Fig 9D-d]. Overall, TOP1 was only found in the cytoplasm when EBOL was present. Since the cytoplasm is where the genomic EBOV RNA is located, there is a stronger correlation to TOP1 playing a role in transcription and replication.
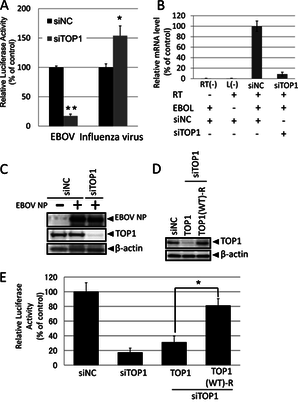
A) Effect of TOP1-knockdown on EBOV polymerase activity.
B) qPCR results of mRNA synthesis in cells treated with siRNA against TOP.
C) Western blot to identify the effect of siTOP1 on viral NP protein synthesis.
D) Western blot confirming the TOP(WT)-R plasmid's success in expressing TOP1 in presence of siTOP1.
E) Identification of TOP(WT)-R's effect on polymerase activity with siTOP1 expression.
Image source: http://jvi.asm.org/content/87/16/8862.full [13]
Further experiments were run to verify whether or not TOP1 had a role in transcription or replication. Short interfering RNAs (siRNAs) targeting TOP1 were used to knockdown the gene. In Fig. 8, they investigated how knocking down TOP1 impacted the polymerase activity, mRNA synthesis, and. protein production. In EBOV, the polymerase activity was reduced by 80%, while siTOP1 enhanced polymerase activity in influenza by 50% [Fig 10A]. The knockdown of TOP1 also resulted in a significant reduction in mRNA synthesis of EBOV genome [Fig 10B]. The presence of siTOP1 did not have an effect on the expression level of the viral protein, NP [Fig 10C]. The lack of an effect on the protein expression indicates that knockdown of TOP1 specifically reduces EBOV polymerase activity. In order to confirm this, a plasmid with RNA interference-resistant TOP1 (TOP(WT)-R) was constructed and introduced into siTOP1. A Western blot was run to verify that TOP1 was expressed in TOP(WT)-R in siTOP1 treated cells [Fig 10D]. After confirming the expression, the plasmid was used to detect polymerase activity in siTOP1 treated cells. TOP(WT)-R was able to successfully restore the polymerase activity [Fig 10E]. This result confirms that TOP1 plays a role in polymerase activity, which in turn is connected to transcription and replication of EBOV.
From the study, Takahashi, et al, were able to confirm that TOP1 plays a significant role in the polymerase activity of EBOV through its interaction with the EBOL. Further experimentation is needed to establish a firm understanding of EBOV transcription and replication. A stronger understanding could potentially lead to development of drugs to treat EBOV.
Conclusion
EBOV is an infectious virus with a limited genome that codes for only seven functional proteins. However, the manner in which EBOV utilizes the seven proteins is quite remarkable. In the sections above, we identified a different protein for each host factor interaction that was utilized to give the virus an increase in infectivity. GP utilized HOPS and NPC1 to assist in the viral entry and viral budding. VP40 and VP24 increased the presence of HSPA5 in order to increase the viral transcript expression to produce more viral proteins. VP24 inhibited STAT1 function to prevent interferons from being produced. L-protein interacts with TOP1 to further progress EBOV genome transcription and replication. Studying host factors can establish a stronger understanding of how EBOV functions and may lead to development of drugs and treatments that will hinder its ability to spread as rampantly as it has.
References
[2] Centers for Disease Control and Prevention. 2014. Ebola Virus Disease Prevention.
[4] Slonczewski, J.L. and Foster, J.W. Microbiology: An Evolving Science. "W.W. Norton & Company, Inc." 2013. Third Ed.
Edited by Amanda He of Joan Slonczewski for BIOL 375 Virology, 2014, Kenyon College.
