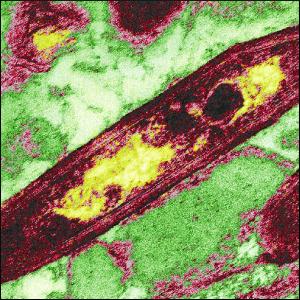
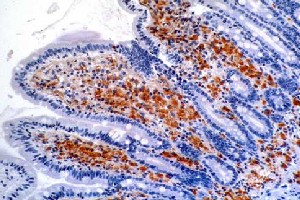
Tropheryma whipplei
Classification
Higher order taxa
Cellular Organisms; Bacteria; Actinobacteria; Actinobacteria (class); Actinobacteridae; Actinomycetales; Micrococcineae; Cellulomonadaceae; Tropheryma; Tropheryma Whipplei (1).
Genus
Genus: Tropheryma
Species: whipplei
Description and significance
Tropheryma whipplei is rod shaped, a gram-postive bacterium (5). It was finally isolated in eukaryotic cells in the year 2000 and propagated in culture at thirty-seven degrees Celsius, but was believed to resist culturing for a long time (3). In fact, it still can only be cultured if part of its eukaryotic host is present. The isolation from human cells and the culturing has allowed more characterization, including sequencing its genome that has resulted in 808 predicted protein-coding gene sequences, even though it grows at a very slow rate of about four to seventeen days. It was first described by the pathologist George H. Whipple in 1907. Still not much is known about the biology of this bacterium, but it wasn't even believed to be a pathogen until it was studied using electron microphotographs of affected tissues in 1961, which was when it was discovered that indeed it is (5). It has a 0.92-Mb genome, placing it in a high-G+C-content gram-positive bacterial group in comparison with others, such as Actinobacteria that have lower G+C contents. Because it is a reduced genomic species, it does not have many genes regulating transcription. This lack of regulation is believed to be the case because inside the host cells the environment is apparently rather stable, and so T. whipplei does not need extensive regulation (3). The way it infects its host and the nautral habitat from which T. whipplie arises is not very well understood as of yet, but it is known to need to live in an aerobic environment that is host-associated (5). However, because it lacks most regulatory components, it seems apparent that this environmental agent is capable of adapting to a wide range of stressful conditions, especially in relation to temperature (3).
It is mesophilic, meaning it grows best at moderate temperatures ranging between twenty-five and forty degrees Celsius. Its optimal temperature is thirty-seven degrees Celsius (5). However, Tropheryma whipplei has a special ability as even though it is not affected by heat shock, it can modify its transcriptome following cold shock at a temperature of four degrees Celsius; this proves that although it lacks a lot of typical regulatory elements, it still has a highly adaptive response to thermal stresses that would be typical with its potential environmental origin being that of something probably at lower temperatures (something still being studied), thereby allowing it to live and adapt in cold conditions (3). It is beneficial for T. whipplei to be able to adapt to a varying range of temperatures because it has no motility (5), but because it has the great ability to adapt during cold shock that is the reason why it is being studied, as well as the fact that it is the pathogen that causes the fatal Whipple diease, as described below within the pathogen section.
Genome structure
It is important for Tropheryma whipplei to have its genome sequenced, because with a 0.92 Mb genome (3), it is the only known human pathogen with a reduced genome sequence within the class of Actinobacteria. This means that this pathogen is the smallest known within its class. It has a circular genome of 927,303 base pairs. T. whipplei has a low G+C content at 46%. Yet, it was originally considered high because 552/808 identified Open Reading Frames have their closest homologues within other actinobacteria class genomes (8). An ORF is important to determine once a gene has been sequenced, as it deals with where the encoded proteins are first transcribed into messenger RNA and then translated into a protein. A particular nucleotide will start and another will stop (known as a stop codon) translation; this is an ORF. Due to its small genomic size, much has yet to be discovered.
Cell structure and metabolism
The most interesting thing about Tropheryma whipplei is the fact that it has such a tiny genome. "Dr Stephen Bentley, who led the team at The Wellcome Trust Sanger Institute, said, 'This really is a wolf in sheep's clothing. Within this amazingly small genome, it has packed a sophisticated array of tools to escape our defense mechanisms. It's an incredibly adept operator which can tell us a great deal about bacteria and their evolution'" (9). It is the "master of disguise", as even though it has only caused a few cases of disease and, once again, has such a small genome, it really molds itself to its environment in a noteworthy way. Just to put into perspective how small it is, most bacterial genomes are three to four times larger, code for about 3,000 genes as opposed to around 800, and have hardly any repetitive DNA. T. whipplei has about five percent of its DNA sequences repeated which also aides in its ability to deviate itself over and over again. It "carries a set of DNA sequences unlike anything previously seen in bacterial genomes." This is the case because it, as far as research has come to discover, selects new regions that it then incorporates into the genes that encodes its single outer coating; this is the reason why Dr. Stephen is quoted saying it is like a wolf in sheep's clothing, because it never gives its host a chance to realize what it is, as it is constantly morphing and changing into something repeatedly different. Not only does it disguise itself in this manner, but it also takes the membranes from the host cells to "hide" in as well. Hence, with these two forms of camouflage, the body doesn't or has a hard time detecting the presence of T. whipplei, as it hides itself in the lining of the human gut, where it gains its energy (9).
Even though it is so small, it still is well equipped for its metabolic activities. However, there are deficiencies of ten amino acids in the biosynthetic pathways. For example, there is a lack of thioredoxin and thioredoxin reductase gene homologues (8). Thioredoxin is usually in its reduced state in an NADPH-dependent reaction, acting as an electron donor. T. whipplei also has a mutation in the DNA gyrase, which relates to the fact that it is resistant to quinolone antibiotic (8). Because of its metabolic deficiencies, like the major energy producing system, which is the Kreb's or TCA it must get the nutrients it needs from the environment, in this case the stomach lining. It also has to rely on its host for the synthesis of essential amino acids like arginine and histidine, as it cannot produce these for itself either(9).
Ecology
Tropheryma whipplei is suspected to have an environmental origin (3), because its closest known relatives originated from the soil; yet, T. whipplei actually is solely dependent on humans. "For nearly 100 years this organism and its host relationship have remained poorly understood." The one factor that is very well understood is that T. whipplei does a good job of using its host without the host ever knowing since it is constantly reshaping itself as previously explained. The fact that it is also very difficult to cultivate without human cells makes understanding this pathogen that much harder(9). All effects it has on its environment and interactions it has with its environment are explained further on in the pathology section, due to the fact that its environment is the human body.
Pathology
Tropheryma whipplei is indeed a pathogen that causes disease in human beings. This not very well-understood pathogen is responsible for causing Whipple disease. This disease is destructive to many of the bodies systems. It causes intestinal malabsorption that leads to cachexia (3). Cachexia is a physical wasting away of the body, and is often caused by disease such as advanced cancer, AIDS, and also in the case of Whipple disease. This wasting syndrome leads to a loss of weight, fat, and muscle, as in healthy people the body adjusts to lose its nutrients much more gradually during a state of starvation, but in the case of cachexia the body will not make any such necessary adjustments. Unfortunately, Whipple disease also effects not only the digestive system, but the cardiac and central nervous systems as well(3). If left untreated, this pathogen is fatal. Yet, even if antibiotic treatment is given, clinical relapses in the form of cerebral issues can ensue. Symptoms include diarrhea and abdominal pain(5), joint pain, arthritis, and even dementia. Due to its ability to constantly change, it manifests in the body in various forms. It can go undetected for decades and then suddenly, without any type of warning, cause a fatal illness. Because it can live inside the body without producing an outward symptoms, it is hard to understand how it causes disease so suddenly after such a prolonged amount of time. Strangely enough, men are eight times more likely to get the disease in comparison to women, typically affecting white, middle-aged men. Between America and Europe, Whipples disease seems to affect people at roughly the same rate, but is much less common in other countries, such as Africa and Asia (7).
Application to Biotechnology
Because the workings and effects of T. whipplei are still being studied, and although it is included in the world of Bioinformatics in the sense that its genomic sequences can be BLASTed, etc., T. whipplei has not been applied to the world of biotechnology in any significant way. Perhaps, once researchers understand more of what it has the potential of producing as far as compounds and enzymes are concerned, or, much more importantly, how it exactly goes about disguising itself so well in the human body, many then it will make a major breakthrough in this field.
Current Research
1. The point of this experiment was to try and find a way to better detect and culture T. whipplei strains. This is due to the fact that molecular-based techniques are less specific and so by taking these pathogens from infected intestinal biopsies would be much better in terms of analysis. The major plus from this experiment was that isolation of new strains of T.whipplei were discovered,and this may end up being a useful tool for diagnosing follow-up patients who suffer with Whipple disease (4).
2. T. whipplei has recently been isolated, with two isolates fully sequenced. Beforehand, the sequence analysis of the 16S rRNA gene determined whether or not one had Whipple disease. However, the high percentage of bacterium in the saliva of healthy people contrasted the negative results gotten with specific target genes. So, the point of this research was to compare the previously determined primers targeting the 16S rRNA gene with realtime Polymerase Chain Reaction (PCR) with probes used to target a specified repeat genes only found in the T. whipplei genome. The study concluded that non-symptomatic carriers of T. whipplei in saliva may exist; however the prevalence of them is much lower than originally reported. Testing the specificity of the designed primers is an important part in avoiding the wrong results, in this case a false-positive. Therefore, at least two specific genes should be targeted before confirming whether or not the person has Whipple disease (6).
3. Because Whipple disease is an infection that targets various organs in the body and is slow growing, many of its characteristics are poorly understood. In this experiment the antibiotic susceptibility that had been tested using real-time quantitative Polymerase Chain Reaction (PCR) assay was used to study three strains of T. whipplei. The results of this particular experiment were that the co-trimoxazole was effective in vitro, but it was due to the fact that it was with sulfamethoxazole. This goes into the fact that metabolically T. whipplei is not capable of many of the typical things that other bacterium are, so this study emphasized the fact that T. whipplei does not contain the encoding gene for the target for trimethoprin, as is to be expected (2).
References
1. Bischoff, Joe, et al. "Tropheryma whipplei." Taxonomy Browser. 10 Jan. 2007. NCBI. 1 June 2007
<http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/ wwwtax.cgi?mode=Info&id=2039&lvl=3&p=mapview&p=has_linkout&p=blast_url&p=genome_blast&lin=f&keep=1&sr chmode=1&unlock>.
2. Boulos, A. "Molecular evaluation of antibiotic susceptibility of Tropheryma whipplei in axenic
medium." Journal of Antimicrobial Chemotherapy 55.2 (Nov. 2004): 178-181. Oxford Journals. 13
Jan. 2005. 27 May 2007 <http://jac.oxfordjournals.org/cgi/content/full/55/2/178>.
3. Crapoulet, Nicolas, et al. "Global Transcriptome Analysis of Tropheryma whipplei in Response to
Temperature Stresses." Journal of Bacteriology 188.14 (July 2006): 28-39. American Society for
Microbiology. Ed. Jack Kenney. 24 Apr. 2006. PubMed Central. 30 May 2007
<http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1539978>.
4. La Scola, Bernard. "Culture and Immunological Detection of Tropheryma whipplei From the Duodenum of
a Patient With Whipple Disease ." Journal of the American Medical Association (JAMA) 285.8
(Feb. 2001). JAMA and Archives. 28 Feb. 2001. 5 June 2007 <http://jama.ama-assn.org/cgi/
content/abstract/285/8/1039>.
5. Raoult D., et al. "Tropheryma whipplei Twist: a human pathogenic Actinobacteria with a reduced genome.", Genome Res, 2003
Aug;13 (8):1800-9
<http://www.ncbi.nlm.nih.gov/sites/entrez?db=genomeprj&cmd=Retrieve&dopt=Overview&list_uids=95>.
6. Rolain, Jean-Marc, et al. "False positive PCR detection of Tropheryma whipplei in the saliva of healthy
people." BMC Microbiology 7 (May 2007). Bio Med Central Microbiology. 5 June 2007. 2 June 2007
<http://www.biomedcentral.com/1471-2180/7/48>.
7. "Tropheryma whipplei causes Whipples disease and has one of the smallest bacterial genomes."
Bacteria Genomes. 2007. European Bioinformatics Institute. 28 May 2007
<http://www.ebi.ac.uk/2can/genomes/bacteria/Tropheryma_whipplei.html>.
8. "Tropheryma whipplei genome database." Structural and Genomic Information Laboratory (IGS). Ed. H.
Ogata. 27 Feb. 2002. National Center for Scientific Research. 3 June 2007
<http://www.igs.cnrs-mrs.fr/mgdb/Tropheryma/>.
9. Von Herbay, Dr. Axel. "Undercover Infective - Cracking the Code of a Quiet Killer." Wellcome Trust
Sanger Institute (Feb. 2003). Wellcome Trust Sanger Institute. 25 Jan. 2007. Wellcome Trust
Genome Campus. 2 June 2007 <http://www.sanger.ac.uk/Info/Press/2003/030221.shtml>.
Edited by Sophia Gevorkian, student of Rachel Larsen and Kit Pogliano.
KMG