Acinetobacter baumannii: The Emergence of a Dangerous Multidrug-Resistant Pathogen
By: Kerri-Lynn Conrad
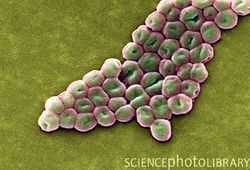
Introduction
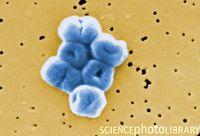
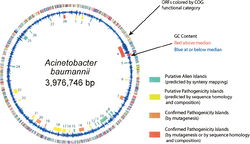
Genome Structure
Cell Structure
Metabolism
Epidemiology

Pathology
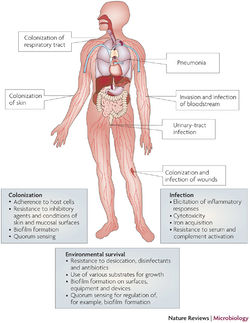
Multidrug-Resistance
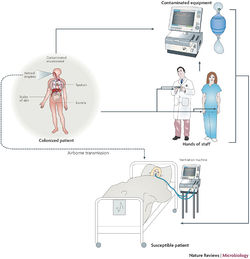
Implications for Hospitals
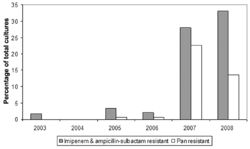
Outbreak in United States Military Treatment Facilities
Implications for Hospitals

Future Work
References
1 Dominguez, G., Dambaugh, T. R., Stamey, F. R., Dewhurst, S., Inoue, N. & Pellett, P. E. 1999. Human herpesvirus 6B genome sequence:
coding content and comparison with human herpesvirus 6A. J Virol 73:8040–8052.
2 Gompels, U. A., Nicholas, J., Lawrence, G., Jones, M., Thomson, B. J., Martin, M. E., Efstathiou, S., Craxton, M. & Macaulay, H. A. 1995. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology 209:29–51.
3 Braun, D. K., Pellett, P. E., Hanson C. A. 1995. Presence and expression of human herpesvirus 6 in peripheral blood mononuclear cells of S100-positive, T cell chronic lymphoproliferative disease. J. Infect. Dis. 171:1351–1355.
4 Meyne, J., Ratliff, R. L., Moyzis, R. K. 1989. Conservation of the human telomere sequence (TTAGGG)n among vertebrates. Proc. Natl. Acad. Sci.USA 86:7049–7053.
5 Clark, D. A. 2000. Human herpesvirus 6. Rev Med Virol 10:155–173
6 Mendez J.C., Dockrell, D.H., Espy, M.J., Smith, T.F., Wilson, J.A., Harmsen, W.S., Ilstrup, D., Paya, C.V. 2001. Human beta-herpesvirus interactions in solid organ transplant recipients. J Infect Dis 183(2):179-184.
7 Nicholas, J., Martin M.E. 1994. Nucleotide sequence analysis of a 38.5-kilobase-pair region of the genome of human herpesvirus 6 encoding human cytomegalovirus immediate-early gene homologs and transactivating functions. J. Virol. 68:597–610
8 Dockrell, D. H. 2003. Human herpesvirus 6: molecular biology and clinical features. J. Med. Microbiol. 52:5–18.
9 Zou, P., Isegawa, Y., Nakano, K., Haque, M., Horiguchi, Y., Yamanishi, K. 1999. Human herpesvirus 6 open reading frame U83 encodes a functional chemokine. J Virol 73:5926–5933.
10 Rotola, A., Ravaioli, T., Gonelli, A., Dewhurst, S., Cassai, E. & DiLuca, D. 1998. U94 of human herpesvirus 6 is expressed in latently infected peripheral blood mononuclear cells and blocks viral gene expression in transformed lymphocytes in culture. Proc Natl Acad Sci USA 95:13911–13916.
11 Chou, S., Marousek G. I. 1994. Analysis of interstrain variation in a putative immediate-early region of human herpesvirus 6 DNA and definition of variant-specific sequences. Virology 198:370–376.
12 Bolle, L.D., Naesens L., Erik, D.C. 2005. Update on human herpesvirus 6 biology, clinical features, and therapy, Clin Microbiol Rev 18:217–245.
13 Brown, N. A., Sumaya, C. V., Liu, C.-R., Ench, Y., Kovacs, A., Coronesi, M., Kaplan, M. H. 1988. Fall in human herpesvirus 6 seropositivity with age. Lancet ii, 396.
14 Ranger S., Patillaud S., Denis F., Himmich A., Sangare A., M'Boup S. 1991.Seroepidemiology of human herpesvirus-6 in pregnant women from different parts of the world. J Med Virol 34:194-8.
15 Enders, G., Biber, M., Meyer, G., Helftenbein, E. 1990. Prevalence of antibodies to human herpesvirus 6 in different age groups, in children with exanthema subitum, other acute exanthematous childhood diseases, Kawasaki syndrome, and acute infections with other herpesviruses and HIV. Infection 18:12–15.
16 Wang, F. Z., Dahl, H., Ljungman, P., Linde, A. 1999. Lymphoproliferative responses to human herpesvirus-6 variant A and variant B in healthy adults. J. Med. Virol. 57:134–139.
17 Collot, S., Petit, B., Bordessoule, D., Alain, S., Touati, M., Denis, F., Ranger-Rogez, S. 2002. Real-time PCR for quantification of human herpesvirus 6 DNA from lymph nodes and saliva. J. Clin. Microbiol. 40:2445–2451.
18 van Loon, N. M., Gummuluru, S., Sherwood, D. J., Marentes, R., Hall, C. B. & Dewhurst, S. 1995. Direct sequence analysis of human herpesvirus 6 (HHV-6) sequences from infants and comparison of HHV-6 sequences from mother/infant pairs. Clin Infect Dis 21:1017–1019.
19 Portolani M., Cermelli C., Moroni A. 1993. Human herpesvirus-6 infections in infants admitted to hospital. J. Med.Virol. 39:146–51
20 Pruksananonda, P., Hall, C.B., Insel, R.A., McIntyre, K., Pellett, P.E., Long, C. E., Schnabel, K. C., Pincus, P. H., Stamey, F. R., Dambaugh, T. R. 1992. Primary human herpesvirus 6 infection in young children. N. Engl. J. Med. 326:1445–1450.
21 Stoeckle M.Y. The spectrum of human herpesvirus 6 infection: from roseola infantum to adult disease. Annu Rev Med 2000: 51: 423–430.
22 Di Luca, D., Mirandola, P., Ravaioli, T., Bigoni, B., Cassai, E. Distribution of HHV-6 variants in human tissues. Infectious Agents and Disease 1996;5:203-14.
23 Luppi, M., Barozzi, P., Morris, C.M., Merelli, E., Torelli, G. 1998. Integration of human herpesvirus 6 genome in human chromosomes. Lancet 352:1707-8.
24 Lusso, P., Gallo, R.C. 1995. Human herpesvirus 6 in AIDS. Immunol Today 16:67-71.
25 Nacheva, E.P., Ward, K.N., Brazma, D., Virgili, A., Howard, J., Leong, H.N., Clark, D.A. 2008. Human herpesvirus 6 integrates within telomeric regions as evidenced by five different chromosomal sites. Journal of Medical Virology, 80:1952–1958.
26 Hall, C.B., Caserta, M.T., Schnabel, K., Shelley, L.M., Marino, A.S., Carnahan, J.A. 2008. Chromosomal integration of human herpesvirus 6 is the major mode of congenital human herpesvirus 6 infection. Pediatrics 122(3): 513–520.
27 Ward, K.N., Leong, H.N., Nacheva, E.P., Howard, J., Atkinson, C.E., Davies, N.W., Griffiths, P.D., Clark, D.A. 2006. Human herpesvirus 6 chromosomal integration in immunocompetent patients results in high levels of viral DNA in blood, sera, and hair follicles. J Clin Microbiol 44:1571–1574.
28 Gompels, U. A., Macaulay, H.A. 1995. Characterization of human telomeric repeat sequences from human herpesvirus 6 and relationship to replication. J. Gen. Virol. 76:451-458.
29 Arbuckle, J.H., Medveczky, M.M., Luka, J., Hadley, S.H., Luegmayr, A., Ablashi, D., Lund, T.C., Tolar, J., DeMeirleir, K., Montoya, J.G., Komaroff, A.L., Ambros, P.F., Medveczky, P.G. 2010. The latent human herpesvirus-6A genome specifically integrates in telomeres of human chromosomes in vivo and in vitro. Proc Natl Acad Sci USA. 107:5563–5568.
30 Daibata, M., Taguchi, T., Taguchi, H., Miyoshi, I. 1998.Integration of human herpesvirus 6 in a Burkitt's lymphoma cell line. Br J Haematol 102:1307-1313
31 Kishi, M., Harada, H., Takahashi, M., Tanada, A., Hayashi, M., Nonoyama, M., Josephs, S.F., Buchbinder, A., Schachter, F., Ablashi, D., Wong-Staal, F., Salahuddin, S.Z., Gallo, R.C. 1988. A repeat sequence, GGGTTA, is shared by DNA of human herpesvirus 6 and Marek’s disease virus. J. Virol. 62:4824–4827.
32 Tanaka-Taya, K., Sashihara, J., Kurahashi, H., Amo, K., Miyagawa, H., Kondo, K., Okada, S., Yamanishi, K. 2004. Human herpesvirus 6 (HHV-6) is transmitted from parent to child in an integrated form and characterization of cases with chromosomally integrated HHV-6 DNA. J Med Virol 73:465-473.
33 Daibata, M., Taguchi, T., Nemoto, Y., Taguchi, H., Miyoshi, I. Inheritance of chromosomally integrated human herpesvirus 6 DNA. Blood 1999; 94:1545-1549.
34 Ablashi, D.V., Eastman, H.B., Owen, C.B., Roman, M.M., Friedman, J., Zabriskie, J.B., Peterson, D.L., Pearson, G.R., Whitman, J.E. 2003. Cross-reactivity with myelin basic protein and human herpesvirus-6 in multiple sclerosis. Annu Neurol. 53(2): 189–197.
Edited by Kerri-Lynn Conrad, student of Joan Slonczewski for BIOL 238 Microbiology, 2011, Kenyon College.
