Metabolisms of Microorganisms Engineered for Ethanol Production
Introduction
Ever since the industrial revolution the world has been heavily relying on fossil fuel as the primary energy source to support economic growth and industrialization. An empirical research conducted in 2010 revealed that nearly sixty countries had passed the maximum petroleum production levels [1]. As the problem of fossil fuel depletion escalates, development of alternative energy sources such as biofuels gradually attracts worldwide attention. The biological production of ethanol biofuel is based on fermentation in microorganisms. Three types of feedstock are generally used for ethanol production: simple sugars from beet and cane, starch from corn and cereals, and lignocellulose from most plants [2].
General Process
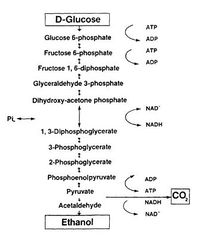
Many microorganisms are capable of converting sugars into ethanol via two steps: glycolysis and fermentation (Fig.1)[2]. During glycolysis, one monosaccharide molecule is converted to two pyruvate at the cost of two NAD+. Under anaerobic condition, the regeneration of NAD+ will be achieved by transferring the electrons from NADH to acetaldehyde (an intermediate formed by the decarboxylation of pyruvate) to form an end-product — ethanol [2].
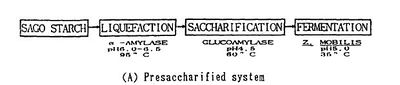
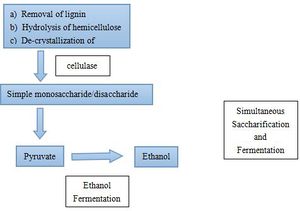
Polysaccharides cannot be directly utilized by microorganism. Thus biological ethanol production using polysaccharides as feedstocks requires additional steps. Starch is an important substrate for ethanol production, but it needs to be hydrolyzed to glucose by liquification and saccharification before ethanol production(Fig.2). Two essential enzymes required for these processes are a-amylase and glucoamylase[3]. Lignocellulose, which is the most abundant plant biomass, is the most common feedstock for ethanol fermentation. Lignocellulose needs to be partially degraded by heating with acid/base catalyst in order for cellulase to interact with the cellulose (Fig.3)[4]. The simple sugars produced from the hydrolysis of cellulose and starch can then be converted to pyruvate by glycolysis by microorganisms. Since saccharification is negatively inhibited by glucose and cellobiose, a method called Simultaneous Saccharification and Fermentation (SSF, combination of cellulose degradation and ethanol fermentation) is commonly used to produce higher ethanol yields in shorter fermentation time [5]. However, SSF requires strict pH and temperature compatibility between the enzymes and the microorganism cultures [4]. Ethanol fermentation pathways in the yeast species Saccharomyces cerevisiae and some Gram-negative bacteria, such as Escherichia coli and Zymomonas mobilis, are the most successful biofuel production process [6].
Saccharomyces cerevisiae
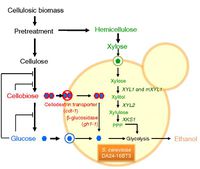
Saccharomyces cerevisiae is an unicellular yeast, capable of utilizing glucose but not xylose as energy source because it lacks xylose reductase(XR) and xylitol dehydrogenase(XDH)[7]. Since xylose is derived from lignocellulose, not being able to ferment xylose is energetically and economically inefficient. To overcome this inefficiency, the incorporation of the genes XYL1 and XYL2, which encode XR and XDH respectively, enables S. cerevisiae to utilize xylose via the pentose phosphate pathway. Despite the acquisition of xylose fermentation pathway, S. Cerevisiae still preferably ferments glucose until all the glucose has been used up. This happens because xylose fermentation is repressed in the presence of extracellular glucose. To minimize the repression by glucose, Suk-Jin (2011) genetically inserted the genes encoding cellodextrin-transporter and β-glucosidase into the genome of S. Cerevisiae[7]. Cellodextrin transporter facilitates the transportation of cellobiose across cell membrane, whereas β-glucosidase catalyzes the hydrolysis of cellobiose into two glucose molecules inside the cell (Fig.4)[7]. This concurrent co-fermentation of glucose and xylose increases ethanol yields.
Escherichia coli
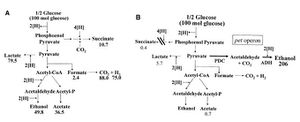
Although the genetically-engineered fermentation pathway in S. cerevisiae has been a great advance in the long journey of bio-fuel development, many environmental factors (e.g. accumulation of ethanol and high salt concentration) can hinder the co-fermentation process. Escherichia coli has been used to avoid these factors. E. coli is able to metabolize a wide spectrum of sugars. E. coli lacks the gene that encodes pyruvate decarboxylase (PDC), instead it expresses pyruvate formate lyase (PFL)[6]. PFL catalyzes the conversion form pyruvate to ethanol at the expense of two NADH per pyruvate used, causing an imbalance of NADH inside the cell (Fig.5A). These imbalance of NADH has to be compensated by the production of acetic and succinic acids, which generates NADH. As a result, sugar fermentation in E. coli yields ethanol and other organic compounds (low ethanol yield)[6]. To bypass this problem, a PET-operon[8], which consists of genes encoding pyruvate decarboxylase and alcohol dehydrogenase, is integrated into the pfl genes inside the genome of E. Coli (Fig.5B). The incorporation of PET-operon, along with the mutations that inhibit succinate production, enables E. coli to yield ethanol exclusively. Since PET operon expression has been found to be correlated to chloramphenicol (Cm) resistance, selecting for Cm-resistant strains of E. coli on acetaldehyde-selection plates ensures high ethanol yields[9]. However, E. coli is also very sensitive to environmental factors such as pH and temperature. Further research is being conducted to improve ethanol production in a more stable manner[6].
Zymomonas mobilis
Zymomonas mobilis is another ethanol-producing microorganism that is capable of fermenting sugars to form ethanol via a distinct mechanism - the Entner-Duodoroff (ED) pathway [10]. Z. mobilis does not possess the enzymes necessary for glycolysis and pentose-phosphate-pathway. Because only one ATP molecule is made through the ED pathway, the growth rate of Z. mobilis is slower. This is one of the advantages of using Z. mobilis to produce ethanol due to the idea that increase in biomass is considered the waste of resources in the context of biofuel production. However, the biggest challenge of using Z. mobilis for ethanol production is that it can only utilize a few simple monosaccharides/disaccharides such as glucose, sucrose and fructose [11]. Genetic engineering has rendered Z. mobilis capable of fermenting cello-oligosaccharides by introducing the gene encoding beta-glucosidase which is attached with a signal peptide derived from glucose–fructose oxidoreductase. The signal peptide facilitates the transportation of beta-glucosidase across the cell membrane[10].
Development of Advanced Biofuels
Ethanol fermentation is one of the most studied topics in the development of bio-fuels, but some disadvantageous molecular properties of ethanol (e.g. High water miscibility and low energy density) limit its application in bio-fuel development[12]. The next generation of bio-fuel begins to emerge as metabolic engineering give insights into building new high-flux pathways for advanced bio-fuel production in microorganisms (Amino acid and acetyl-CoA pathway). Since the construction of the engineered biosynthetic pathways of these advanced biofuels are much more complex than ethanol fermentation, manipulation of building blocks derived from pathways beyond glycolysis is involved [12].
References
[1].Hook M, Tang X. 2013. Depletion of fossil fuels and anthropogenic climate change - A review. Energy Policy. 52: 797-797.
[2].Stewart GG, Panchal CJ, Russel I, Sills AM. 1983. Biology of ethanol-producing microorganisms. Critical Reviews in Biotechnology. 1: 161-188.
[3].Sang KR. 1991. Metabolic engineering of zymomonas mobilis for ethanol production from starch. 한국미생물학회 학술대회논문집. 4: 97-109.
[4].Lin Y, Tanaka S. 2006. Ethanol fermentation from biomass resources: current state and prospects. Applied Microbiology and Biotechnology. 69: 627-642.
[5].Cardona CA, Sánchez ÓJ. 2007. Fuel ethanol production: Process design trends and integration opportunities. Bioresource Technology, 98: 2415-2457.
[6].Dien BS, Cotta MA, Jeffries TW. 2003. Bacteria engineered for fuel ethanol production: Current status. Applied Microbiology and Biotechnology. 63: 258-266.
[7].Ha S, Galazka JM, Kim SR, Choi J, Yang X, Seo J, Yang X, Seo J, Glass NL, Cate JHD, Jin, Y. 2011. Engineered Saccharomyces cerevisiae capable of simultaneous cellobiose and xylose fermentation. Proceedings of the National Academy of Sciences of the United States of America. 108: 504-509.
[8].Tzagoloff A, Dieckmann CL. 1990. PET genes of saccharomyces cerevisiae. Microbiological Reviews. 54: 211-225.
[9].Rao K, Chaudhari V, Varanasi S, Kim D. 2007. Enhanced ethanol fermentation of brewery wastewater using the genetically modified strain E. coli KO11. Applied Microbiology and Biotechnology. 74: 50-60.
[10].Yanase H, Nozaki K, Okamoto, K. 2005. Ethanol production from cellulosic materials by genetically engineered Zymomonas mobilis. Biotechnology Letters. 27: 259-263.
[11].Bai FW, Anderson WA, Moo-Young M. 2008. Ethanol fermentation technologies from sugar and starch feedstocks. Biotechnology Advances. 26: 89-105.
[12].Wen M, Bond-Watts BB, Chang MCY. 2013. Production of advanced biofuels in engineered E. coli. Current Opinion in Chemical Biology. 17: 472-479.