Orthomyxoviridae
A Viral Biorealm page on the family Orthomyxoviridae
Baltimore Classification
Higher order taxa
Viruses; ssRNA negative-strand viruses; Orthomyxoviridae
Genera
Influenza A virus, Influenza B virus, Influenza C virus, Isavirus, Thogotovirus
Description and Significance
The orthomyxovirus gets its name from the Greek word 'myxa' that means mucus.
Influenza virus types A and B are both common causes of acute respiratory illnesses. Both virus types may cause epidemics of considerable morbidity and mortality but influenza B infections are often limited to localized outbreaks whereas influenza A viruses are the principal cause of larger epidemics including worldwide pandemics. Influenza occurs in winter epidemics that affect 1-5% of the population in temparate regions. Influenza can be contracted throughout the year in tropical regions and its contribution to overall morbidity and mortality is less well defined. (source: World Health Organization: Immunization, Vaccines and Biologicals)
Genome Structure
The genome of the orthomyxovirus consists of six segments to eight segments of linear, negative-sense, single-stranded RNA. The complete genome is 10000-14600 nucleotides long. Segment 1 is fully sequenced and the complete sequence is 2300-2500 nucleotides long. Although sequenced and of the same length as Segment 1, segment 2 only has an estimate of the sequence so far. Segment 3 is also sequenced, but only estimated, and the complete sequence is 2200-2300 nucleotides long. Segment 4 has been completely sequenced and the complete sequence is 1700-1800 nucleotides long. Segment 5 has been sequenced, but only estimated, and is 300-1900 nucleotides long. Segment 6 has been sequenced, but only estimated, and is 1400-1500 nucleotides long. Segment 7 has been sequenced, but only estimated, and the complete sequence is 800-1100 nucleotides long. The genome has terminally redundant sequences and the sequence is repeated at both ends. The nucleotide sequences at the 3'-terminus are identical. The 5'-terminal sequence has conserved regions and repeats complementary to the 3'-terminus; terminal repeats at the 5'-end are 11-14 nucleotides long. The 3'-terminus has conserved nucleotide sequences; is 11-13 nucleotides long; in the genera of same family. The sequence has conserved regions in all RNA species or some RNA segments. The multipartite genome is encapsidated with each segment in a separate nucleocapsid, and the nucleocapsids are surrounded by one envelope. Each virion contains defective interfering copies. (source: ICTV dB Descriptions)
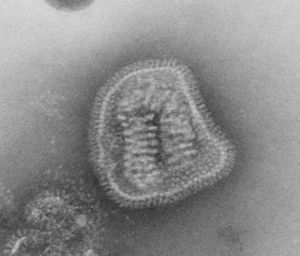
Virion Structure of an Orthomyxovirus
The virions of an orthomyxovirus consist of an envelope, a matrix protein, a nucleoprotein complex, a nucleocapsid, and a polymerase complex. The virus capsid is enveloped. The virions are spherical to pleomorphic and filamentous forms occur. The virions are 80-120 nm in diameter and 200-300(-3000) nm long. The surface projections are densely dispersed distinctive hemagglitinin-esterase (HEF) spikes, or spaced widely apart hemagglutinin (HA) spikes. Clusters of neuramidase (NA) irregularly inerpose the major glycoprotein in a ratio of HA to NA about 4-5 to 1. There are about 500 spikes evenly dispersed or clustered and are covering the surface comprising hemagglutinin, or neuraminidase, or esterase-esterase. The surface projections are composed of one type of protein or different types of proteins and are 10-14 nm long and 4-6 nm in diameter. The nucleocapsid is elongated with helical symmetry and is segmented with loops at one end. The segments have different sized classes with clear predominate lengths with a length of 50-130 nm (in differnent class sizes) and a width of 9-15 nm. (source: ICTV dB Descriptions)
Reproductive Cycle of an Orthomyxovirus in a Host Cell
It takes about 6 hours for the replication of the orthomyxovirus, killing the host cell in the process. The virus attaches to the permissive cells via the hemagglutinin subunit, which binds to cell membrane glycolipids or glycoproteins containing N-acetylneuraminic acid, the receptor for virus adsorption. The virus is then engulfed by pinocytosis into endosomes. The acid environemnt of the endosome causes the virus envelope to fuse with the plasma membrane of the endosome, uncoating the nucleocapsid and releasing it into the cytoplasm. A transmembrane protein derived from the matrix gene (M2) forms an ion channel for protons to enter the virion and destabilize protein binding, allowing the nucleocapsid to be transported to the nucleus, where the genome is transcribed by vital enzymes to yield viral mRNA. Orthomyxovirus replication depends on the presence of active host cell DNA, unlike the replication of other RNA viruses. The virus scavenges cap sequences from the nascent mRNA generated in the nucleus by transcription of the host DNA and attaches them to its own mRNA. These cap sequences allow the viral mRNA to be transported to the cytoplasm, where it is translated by host ribosomes. The nucleocapsid is assembled in the nucleus.
Virions acquire an envelope and undergo maturation as they bud through the host cell membrane. The viral envelope hemagglutinin is subjected to proteolytic cleavage by host enzymes during budding. This process is necessary for the released particles to be infectious. Newly synthesized virions have surface glycoproteins that contain N acetylneuraminic acid as a part of their carbohydrate structure, and this are vulnurable to self-agglutination by the hemagglutinin. A major function of the viral neuraminidase is to remove these residues. (source: Medical Microbiology)
Viral Ecology & Pathology
Influenza virus is transmitted from person to person primarily in droplets released by sneezing and coughing. Some of the inhaled virus lands in the lower respiratory tract, and the primary site of the disease is the tracheobronchial tree, although the nasopharynx is also involved. The neuraminidase of the viral envelope may act on the N-acetylneuraminic acid residues in mucus to produce liquefaction. This liquified mucus may help spread the virus through the respiratory tract in concert with the mucociliary transport. The superficial mucosa suffers cellular destrcution and desquamation because of the infection of the mucosal cells. Nonproductive cough, sore throat and nasal discharge are some sysmptoms that result from the endema and mononuclear cell infiltration of the involved areas. The cough may be persistent but the most prominent symptoms of influenza are systemic-- fever, muscle aches and general prostration. These systemic symptoms are not caused directly by the virus because viremia is rare. A possible cause is circulating interferon, as administration of theraupetic interferon causes systemic symptoms resembling those of influenza.
The evidence so far indicates that the extent of virus-induced cellular destruction is the prime factor determining the occurence, severity and duration of clinical illness. It is possible to recover virus from respiratory secretions for 3 to 8 days in an uncomplicated case. At times of maximal illness, peak quantities of 104 to 107 infectious units/ml are detected. The titer begis to drop in concert with the progressive abatement of disease after 1 to 4 days of peak shedding
The infection may extensively involve the alveoli, particularly in patients with underlying heart or lung disease. This may result in interstitial pneumonia, sometimes with marked accumulation of lung hemorrhage and endema. Pure viral pneumonia of this type is a severe illness with a high mortality. Virus titers in secretions are high, and viral shedding is prolonged. However, in most cases, bacteria is the causative agent of pneumonia associated with influenza. Examples include pneumococci, staphylococci, and Gram-negative bacteria. The preceding viral infection damages the normal defenses of the lung, setting the stage for the bacteria to invade and cause disease. (source: Medical Microbiology)
The Biology of Influenza
H*N* Nomenclature for Influenza A
The H*N*, where “*” is a number, is one of the naming systems for the Influenza A genus. Influenza A virus is the only species in the genus and is broken down into subtypes with this naming system.
“H” – Hemagglutinin
H stands for hemagglutinin which is an antigenic glycoprotein that allows the virus to attach to the cell being infected. It attaches using two types of “chains”. The first chain searches for specific sugar chains on an organism's cellular proteins. Once found, the first chain binds to the cell and the second chain begins the attack on the cell. The protein name derives from its ability to agglutinate red blood cells. The virus coat is covered with hemagglutinin molecules so that a virus particle can bind to many binding red blood cells at once, forming a visible clump (1).
There are 16 subtypes of hemagglutinin, H1-H16, the most recent found in 2004 in black-headed gulls from Sweden (2). H1, H2 and H3 have typically been the subtypes that have been observed in humans.
“N” – Neuraminidase
N stands for neuraminidase, an antigenic glycoprotein enzyme that is embedded in the surface of the influenza virus. Neuraminidase allows the virus to escape the cell after it has replicated. This enzyme cleaves through part of the cell's plasma membrane to extricate itself and move on to another cell. Different levels of virulence are often linked to different subtypes of neuraminidase depending on their activity. The faster the neuraminidase scythes through the plasma membrane the quicker the virus can get out and infect other cells (3).
The numbering system for neuraminidase is by subtype, similar to hemagglutinin, with 9 known subtypes. N1 and N2 have been identified in human pathogenic strains. The rest occur primarily in avian variations of influenza (3).
References
1. http://www.rcsb.org/pdb/static.do?p=education_discussion/molecule_of_the_month/pdb76_1.html>
2. Fouchier, R.A.M., et al. (2005) “Characterization of a Novel Influenza A Virus Hemagglutinin Subtype (H16) Obtained from Black-Headed Gulls” J Virology 79:2814-2822
3. Winquist, A., et al. “Neuraminidase Inhibitors for Treatment of Influenza A and B Infections,” http://www.cdc.gov/mmwr/preview/mmwrhtml/rr4814a1.htm
The “Spanish Flu” of 1918-1919 – The “Great Pandemic”
In 1918, the European powers had been locked in a devastating war for over three years. Millions would die before the warring nations would sign an armistice in November, 1918. Unfortunately, a new strain of influenza was poised to spread throughout the globe at an alarming speed. The world was too preoccupied to focus on this new public health concern, but the 1918 flu would soon take more lives than World War I.
The onset of this new influenza occurred very suddenly and the geographic origin of the disease remains unclear. The outbreak appears to have begun in the United States, who had only entered the war the previous year. Some of the earliest reported cases occurred in American military bases, such as Camp Funston, Kansas. US soldiers spread the disease to Europe, where it spread across the entire continent. The first wave of influenza was known as the “three day fever.” Victims recovered within days, and the mortality rate was very low. The exception was Spain, which had eight million cases reported in May, 1918. The virus in Spain affected nearly half the total population of twenty million people and resulted in 255,000 deaths. Due to Spain’s lack of wartime censorship, most of the world became aware of the disease through reports from Spain, hence the nickname of “Spanish flu.”
A more lethal second wave of influenza emerged in August, 1918 and spread across the globe by November. The mass movement of people that occurred during the war helped the disease spread quickly, even to remote regions. These first two outbreaks were unusual because they occurred outside of the typical flu season, leading some to speculate that the Germans were using biological warfare, even though both sides were affected. In the closing days of the war, many men were too sick to continue fighting. Due to the shortage of healthy soldiers, battle strategies were changed and several fleets were left unmanned. The large public gatherings on Armistice Day also facilitated the spread of the disease. The severity of the outbreak tested the limits of contemporary healthcare.
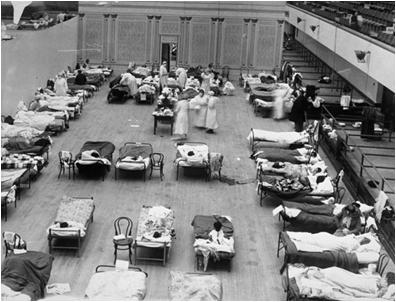
Red Cross nurses care for flu patients in Oakland Municipal Auditorium, 1918 (Public domain image from Wikipedia) - http://en.wikipedia.org/wiki/Spanish_flu
The American health care system was unprepared for a serious epidemic. Because of the influx of injured soldiers returning from Europe, hospitals were not staffed or equipped to handle an epidemic. Many doctors fell ill, creating a further shortage of medical personnel. The Red Cross established the National Committee of Influenza to properly utilize resources. Both state and federal governments also stepped in to help. Ordinances were put in place to curb the disease’s spread. Gauze masks were distributed, certificates of health were required for railroad travel, and funerals were limited to fifteen minutes. The American Public Health Association even attempted to ban public coughing and sneezing. These rules were generally accepted both because of wartime and because of increased government authority during World War I.
In early 1919, a third wave of influenza struck. Never before had successive flu epidemics occurred so rapidly and with such a high mortality rate. Unlike other flu outbreaks, adults between the ages of 20 and 40 had the highest death rate for all three waves. Mortality rates for 15-34 year olds were twenty times higher than in previous flu epidemics, and 99% of all deaths were in people under 65 which contrast sharply with most influenza deaths.
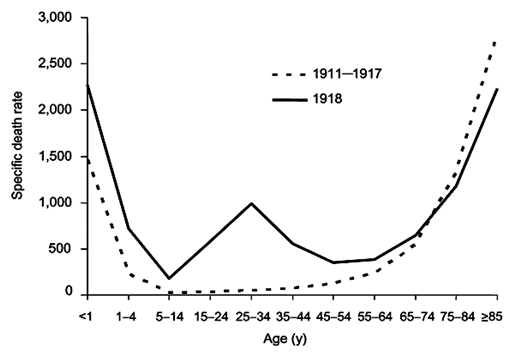
Normal flu mortality rate compared to 1918 outbreak. (Public domain image from Wikipedia - http://en.wikipedia.org/wiki/Spanish_flu)
Death usually came very fast for all age groups. Patients would develop extreme cases of pneumonia and die within 3-5 days. In some cases patients’ lungs would fill with fluid mere hours after showing early symptoms. In contrast, normal flu symptoms of sore throat, fever and chills typically last a few days before the patient recovers.
A quarter of America was infected, resulting in 550,000-675,000 deaths. Half of American casualties in World War I were from influenza, not combat. The average American lifespan declined by 10-12 years during this period. Major eastern cities were hit the hardest because so many soldiers passed through their ports. In the fall of 1918, Philadelphia had 13,000 casualties in only three weeks. Boston also faced a severe outbreak, and its Commonwealth Pier became notorious for its role in spreading the disease.
Estimates vary, but anywhere from 25-50 million people died from Spanish flu worldwide. Some sources suggest 100 million deaths. A fifth of the world was infected. In India, 12-17 million died. By comparison, approximately 16 million people died in all of World War I. The global mortality rate is estimated to have been around 2.5%, compared to less than 0.1% for most flu outbreaks. Spanish Flu is probably responsible for more total deaths in a short period of time than any pandemic in history, including the Black Plague.
Unusual Fatality Distribution—The Cytokine Storm
An unusual feature of the 1918 pandemic was who it killed. Most flu fatalities are among young children and the elderly. The 1918 virus killed mostly healthy adults between the ages of fifteen and forty-five—people with generally robust immune systems. It is believed that it was their robust immune systems that killed them. They would have likely been exposed to other H1N1 viruses in childhood, enabling faster recognition by the immune system. Infection by the 1918 flu virus produced a “cytokine storm.”
A cytokine is a generic term for regulatory molecules (proteins or glycoproteins) produced by various immune cells in the course of a response. A cytokine storm results from an excessive immune system reaction that damages the body, causing failure of multiple organ systems. Cytokine storms occur very rarely, which makes them exceptionally difficult to study in-depth. Such over-reaction typically leads to acute respiratory distress syndrome, an often fatal condition that causes severe damage to alveoli and lung tissue. The cytokines released cause vasodilation and increased vascular permeability. Thus, the capillaries in the lungs become “leaky,” causing the lung to fill with fluid and essentially drown the victim. The primary cause of death is not lung failure but rather multi-system organ failure due to lack of oxygen. The fluid also provides a medium for bacterial growth, encouraging pneumonia. The reason that the 1918 influenza virus was so dangerous to young adults was, almost paradoxically, apparently because of their stronger immune system. A stronger immune system generally produces a stronger response and a greater overreaction. Children and the elderly have less robust immune systems and as a result had much weaker cytokine storms, hence the inversion of the usual pattern of mortality.
Reconstruction of the 1918 H1N1 virus
In 1995, work on reconstructing the 1918 H1N1 virus began. Researchers obtained RNA fragments from preserved tissue samples and the cadaver of a victim found buried in the Alaskan permafrost. Using these RNA sequences, cloned segments of DNA were produced and introduced into cells. The sequencing of the genome was completed in 2005.
The 1918 virus is believed to have originated entirely in birds (as opposed to co-infecting a host along with a human virus), although there are elements in the viral genome that do not appear in avian viruses. The “rescued” 1918 virus was used to infect mice. The mice lost thirteen percent of their body weight in two days, an indication of the virulence of the 1918 flu.
Most of the victims of the 1918 pandemic died from pneumonia. By inserting genes from the 1918 virus into a modern viral strain, three genes were implicated in this effect. Normally, influenza infects only the upper respiratory tract of its host. The PA, PB1 and PB2 as well as a nucleoprotein from the 1918 virus allowed common modern flu viruses to multiply deep in the lung tissue of lab ferrets, who died of pneumonia.
Responses to the Spanish Flu
The Spanish Flu affected many countries. Across the entire globe, the number of deaths was estimated to be as high as 100 million. The flu hampered military operations during the last year of World War 1: Germany, Turkey, Austria-Hungary, and France all suffered major casualties among troops and the civilian population. Every country reacted differently to the disease and its effects on the population.
Though the virus was confined to military camps for most of the summer of 1918, the flu reached Philadelphia by September. The government ordered mandatory vaccinations and all influenza cases were to be reported to the health department. The flu began to spread rapidly through the country, causing the police to close schools, churches, and other public places in order to quarantine the population. The quick spread of the epidemic gave little time for preventive measures to be taken. Scientific studies of prevention plans show that San Francisco, Milwaukee, Kansas City, and St. Louis had the most effective responses. St. Louis actually began its prevention plan two weeks before Philadelphia, the hardest hit city initially.
In Canada, the influenza pandemic had a positive impact on Canada’s public health system. The Canadian Medical Association had been pushing for federal health regulations since 1900. In 1919, the Department of Health Act was passed, and funds were finally available to expand hospitals and health care. The influenza epidemic also forced officials to cancel the Stanley Cup Playoffs. With the series between the Montreal Canadiens and the Seattle Metropolitans tied, several players from Montreal became severely ill with the flu, and one later died. The remainder of the series was cancelled.
A worldwide trend of declining pregnancies and live births was generally seen in the amount of during the early 1920s that can largely be attributed to influenza deaths. (This contrasts with the “baby boom” after World War II). Pregnant women appeared more susceptible. About 20-50% of pregnant women struck by the flu died during the epidemic. In most European countries, the total fertility rate dropped 7%, with Norway being an exception showing an increase in birth rate.
Treatment of the Spanish Flu
The Spanish Flu was very virulent, and quick to kill. With so many people dying, treatments had to be found as quickly as possible. Unfortunately, there were few effective means of combating the disease. Out of desperation, doctors even tried bleeding patients, a long discredited and abandoned practice, but to no effect. Oxygen was also given to those who were ill without success. Doctors and researchers tried to develop new vaccines as well. With any strain of flu, it is difficult to make a vaccine and none of them appeared to have significant positive effect. Despite their efforts, medical personnel found that the only thing that could be done was to allow the disease to run its course, giving plenty of fluids and allowing the patient to rest. This is still the treatment for most cases of flu.
Other than rest and fluids, blood transfusions seemed to be the only truly effective form of treatment. Transfusions of whole blood, plasma, or serum from survivors of the flu were given to people who were very ill. The treatment was effective because the survivors had developed antibodies which were transferred with their blood. Even this treatment was not completely effective, though it appeared to reduce mortality rates by up to 50%. It is unclear how effective transfusion would be as a treatment for a modern flu epidemic.
Historical Remembrance and Contemporary Relevance
The 1918 influenza pandemic holds a unique place in the history of epidemics. Compared to other influenza epidemics, it had death rates 5 – 20 times higher than expected. Between 50 and 100 million people lost their lives, including a surprisingly high number of healthy individuals and an unexpected number of youths, neither of which were the traditional victim groups for influenza.
In America, despite the unusual potency and unique victim groups of the Spanish Flu, it has become the “forgotten epidemic”. Several theories attempt to explain this historical amnesia. The most important explanations include the epidemic’s close proximity to World War I and the relative rapidity of the Spanish Flu, which would move in unexpectedly on communities, wreak havoc, and then disappear shortly afterwards.
Whether Americans remember the 1918 Pandemic or not, the causal H1N1 virus has continued to play a role in history. Indeed, all “Influenza A pandemics since that time…have been caused by descendants of the 1918 virus, including ‘drifted’ H1N1 viruses and reassorted H2N2 and H3N2 viruses…making the 1918 virus indeed the "mother" of all pandemics”.
The contemporary influenza threat, however, comes from the H5N1 avian flu strain (see below), which is only distantly related to H1N1. Though both strands are avian in origin, they appear to have arisen through different evolutionary pathways. Many scholars who believe the world is “overdue” for a cyclical influenza outbreak believe H5N1 is the leading contender. The importance of remembering the Spanish Flu pandemic today may be its usefulness for comparisons to hypothetical modern outbreaks.
Recent studies of the 1918 outbreak in America imply that some actions taken by city authorities, such as quarantines – which were previously thought rather unsuccessful on the whole – and the wearing of masks were effective in mitigating the damage of the epidemic. Problems arose, however, when these sanctions were lifted too soon and the Spanish Flu returned in subsequent waves. In the highly mobile modern world, state-imposed isolation appears a far-fetched and nearly unenforceable intervention. Requiring face masks in public, though more plausible, could also prove difficult to implement in the current individual-based culture. Plus, the original state-intervention measures required a consensus among authorities over the best course of action, quick decision making, and a large amount of public support. In 1918, America was embroiled in the First World War and the citizenry was largely willing to allow extra governmental power in hopes of ending the epidemic. In contrast, the citizenry today seems far less likely to grant additional authority to the state. Regardless of these non-pharmaceutical interventions, doctors seem to be in agreement that a vaccine would still be the primary focus of health authorities if an avian flu epidemic were to strike.
References
“Avian Influenza or Bird Flu: Reference Summary.” (2008) “X-Plain Avian Influenza or Bird Flu,” http://www.nlm.nih.gov/medlineplus/tutorials/avianflu/id509103.pdf
Bakalar, N. (2007) “How (and How Not) to Battle Flu: A Tale of 23 Cities” New York Times http://www.nytimes.com/2007/04/17/health/17flu.html
Baum, L. (2006) “The Deadliest Fall,” http://ideaexplore.net/news/041116.html
Billings, M. (2005) “The Influenza Pandemic of 1918.” http://www.stanford.edu/group/virus/uda/
Borenstein, S. (2007) “Research on monkeys finds resurrected 1918 flu killed by turning the body against itself,” http://www.usatoday.com/tech/science/discoveries/flu-research.htm?POE=NEWISVA
Center for Infectious Disease Research and Policy, University of Minnesota, http://id_center.apic.org/cidrap/content/influenza/panflu/biofacts/panflu.html
Cummings, S. “Spanish Influenza Outbreak, 1918” http://history-world.org/spanish_influenza_of_1918.htm
Duffy, M. (2002) “The Influenza Pandemic” First World War: The War to End All Wars. http://www.firstworldwar.com/atoz/influenza.htm
"Influenza Epidemic of 1918–19" (2009) Encyclopædia Britannica Online http://www.britannica.com/EBchecked/topic/287805/influenza-epidemic-of-1918-19
Kindt, T.J., R.A. Goldsby and B.A. Osborne (2007) Kuby Immunology, 6th Revised Ed, WH Freeman and Co, NY; p 302
Lamb, R.A, and D. Jackson (2005) “Extinct 1918 virus comes alive,” Nature Med 111154 – 1156
Mamelund, S.-E. (2001) “Effects of the Spanish Influenza pandemic on fertility and nuptiality in Norway” http://www.iussp.org/Brazil2001/s30/S34_P01_Mamelund.pdf
Morens, D.M., and A.S. Fauci (2007) “The 1918 Influenza Pandemic: Insights for the 21st Century,” J Infect Dis 195:1018-1028
Parsons, D. (2006) “The Spanish Lady and the Newfoundland Regiment” WWI: The Medical Front. Dr. Geoffrey Miller” http://www.vlib.us/medical/parsons.htm
Osterholm, M.T. (2005) “Preparing for the Next Pandemic,” New Eng J Med 352:1839-1842
Osterholm; M.T., and A.L. Petrosino (2005) “Cytokine Storm and the Influenza Pandemic,” http://www.cytokinestorm.com/
Researchers unlock secrets of 1918 flu pandemic” (2008) http://www.reuters.com/article/newsOne/idUSTRE4BS56420081229?pageNumber=1&virtualBrandChannel=0
Schoenstadt, A. (2008) “Spanish Flu” http://flu.emedtv.com/spanish-flu/spanish-flu.html
Taubenberger, JK., and D.M. Morens “1918 Influenza: The Mother of All Pandemics.” The Center for Disease Control. 2005. Emerging Infectious Diseases. 8 April 2009. <http://www.cdc.gov/ncidod/eid/vol12no01/05-0979.htm#cit>.
“The Great Pandemic: The United States in 1918 – 1919.” http://1918.pandemicflu.gov/the_pandemic/04.htm
“The Threat of Pandemic Influenza: Are We Ready?” National Academies of Science Workshop Summary (2005) http://www.nap.edu/openbook.php?record_id=11150&page=1
“The Deadly Virus: The Influenza Epidemic of 1918” National Archives and Records Administration http://www.archives.gov/exhibits/influenza-epidemic/
“The 1918 Flu Virus is Resurrected” (2005) Nature 437:794-795.
“1918 Spanish flu treatment may also be effective for current avian influenza patients,” http://www.news-medical.net/?id=19832
Other Influenza Epidemics
The Asian Flu 1957-1958
The 1957-1958 Asian flu pandemic was not as virulent as the 1918 pandemic. There were far fewer deaths and cases of the flu, but it was important for two reasons. First, the incidence rate was highest for children; second, it was the first pandemic for which a vaccine was available. The pandemic was responsible for an estimated 2 million deaths worldwide. Those most affected by the pandemic were school children, young adults, pregnant women, and the elderly
The pandemic had two waves. It began in China in February 1957. By June it had spread to Asia, Europe, and America, affecting primarily school age children. A second wave occurred in January and February, 1958 and affected the elderly more than children. The first wave was attributed to children going back to school in the Fall of 1957. The close contact with other children helped spread the disease. The average age of those affected was 6-12. Schoolchildren then helped spread the flu by taking it home to their families. Many schools closed for a time to combat the spread of the pandemic, a measure that was somewhat effective. Although the infection rate in the first wave was highest for children and adolescents, the elderly had the highest death rate. The lower incidence in the elderly may have been due to partial immunity because most had been exposed to the Spanish flu of 1918. They had a higher death rate however because of their age.
Because of medical advances since the 1918 Spanish flu, the virus responsible for this new epidemic was quickly identified as a H2N2 type, and a vaccine was quickly made. This was the first vaccine available for a flu pandemic. It was available by May of 1957, it became generally accessible to the U.S. by August and by October in Britain. Despite availability, in the U.S., less than half of the 60 million doses were used.
References:
http://cns.miis.edu/flu_watch/history.htm
“Influenza: A Short History of the Disease,” http://lhncbc.nlm.nih.gov/apdb/phsHistory/health_news.html
“Pandemic Influenza,” http://www.globalsecurity.org/security/ops/hsc-scen-3_pandemic-influenza.htm
“1957: British public gets 'Asian Flu' vaccine,” http://news.bbc.co.uk/onthisday/hi/dates/stories/october/1/newsid_3086000/3086843.stm
The Hong Kong Flu 1968-1969
The Hong Kong virus, an H3N2 strain, evolved from the Asian Flu virus, which circulated from 1957 to 1968. (3, 7) The Hong Kong flu was probably a result of antigenic drift, meaning that the original virus was an Asian Flu virus with an advantageous mutation that kept the human immune system from recognizing it as a pathogen. Because Hong Kong Flu evolved from Asian Flu, many people who had Asian Flu were immune or partially immune, largely due to the overlapping N2 subtype. Mortality worldwide was 500,000 to 750,000 from 1968 to 1969. The United States recorded 34,000 deaths. The Hong Kong Flu reappeared in 1970 and in 1972 but has not been seen since.
Hong Kong Flu appeared in the United States in September of 1968, possibly brought by soldiers returning from Vietnam. The incidence escalated through December and into January, the month with the highest number of fatalities. This is a slightly atypical timeline. Flu season typically begins in early October and goes through March, with the highest number of cases in occurring in February. The Hong Kong Flu peaked several weeks earlier. The height of the infectious period coincided with schools’ winter breaks. Children, who suffered the highest incidence of infection, were at home where they were more isolated than if they had been at school. This may have lowered the number of people exposed to contagious patients and stunted the flu’s spread. Many schools closed which likely also helped keep the pandemic from worsening. The US Department of Health and Human Services called it “the mildest pandemic of the twentieth century.”
Hong Kong Flu was first recognized in July of 1968 and the World Health Organization (WHO) determined its H3N2 type on August 16th. The WHO provided antibiotics, though as is always the case with a viral infection, they were ineffective. Vaccines were manufactured in the United States, but they were not finished in time to prevent the disease from spreading.
References
http://medicine.science-tips.org/health/diseases-and-conditions/hong-kong-flu.html
http://cns.miis.edu/flu_watch/history.htm
“Pandemics and Pandemic Scares in the Twentieth Century,” http://www.hhs.gov/nvpo/pandemics/flu3.htm
Earn, D.J.D., J. Dushoff and S.A. Levin (2002) “Ecology and evolution of the flu,” Trends Ecol Evol 17:334-340.
http://www.medterms.com/script/main/art.asp?articlekey=26429
Harder, T.C., and W. Ortrud (2006) “Avian Influenza,” in Influenza Report, ed. B.S. Kamps, C. Hoffmann and W.Preiser, Flying Publisher (www.flyingpublisher.com), Ch. 2
http://www.cdc.gov/flu/about/season/flu-season.htm
http://www.pandemicflu.gov/general/historicaloverview.html
http://www.publications.parliament.uk/pa/ld200506/ldselect/ldsctech/88/8805.htm
Avian Influenza A, H5N1
Introduction
Avian influenza A (H5N1) is an influenza virus subtype that occurs primarily in bird populations. The current form of virus does not typically infect humans unless there is direct, extensive contact with infected poultry or other contaminated surfaces (1). Fewer than 500 human cases of H5N1 influenza have been reported worldwide in the 5 years since the virus first crossed the species barrier from birds to humans. Nonetheless, there is cause for concern because the H5N1 virus has caused the largest number of cases of severe disease and death in humans of any avian flu virus. At present, human-to-human spread of the virus has been “limited, inefficient and unsustained” (2). Even without human-to-human transmission, fatality rates are over 50%, far higher than from any other flu epidemic in history (2,3). By comparison, the fatality rate for recent pandemics has been about 0.1% while the Spanish flu of 1918-1919 had a fatality rate of about 2.5%. This high rate for the H5N1 virus is presumably because there is little preexisting immunity to its (4). Should the virus develop an improved ability to spread from person-to-person, a devastating pandemic could occur. It should be noted that the fatality rate data collected by the WHO may be skewed because it is the most severe cases that are reported. Cases that are not as severe go unreported to the WHO, thus making it seem as if there are a disproportionate number of fatalities (5).
History
The highly pathogenic H5N1 virus was first isolated in 1996 from farmed geese in Guangdong Province, China near Hong Kong. T he following year, the first confirmed cases of human infection with H5N1 were reported in 18 patients in Hong Kong with 6 fatalities (6). By 2004, the virus had spread through the bird populations of East Asia, becoming endemic. Widespread human cases were reported for the first time.
The virus has predominantly affected humans in Vietnam and Indonesia and China with fewer cases in other Southeast Asian countries (see Table). Egypt has also shown a number of cases with a very high fatality rate. Over the last several years the H5N1 virus has become increasingly endemic in bird populations further and further from Hong Kong. Infections in birds have been confirmed throughout most of Asia and the Middle East, with human cases reported as well in countries as far from Hong Kong as Nigeria (2).
Human Incidence of H5N1 by Country (2)
| Country | Cases | Deaths |
| Azerbaijan | 8 | 5 |
| Bangladesh | 1 | 0 |
| Cambodia | 8 | 7 |
| China | 38 | 25 |
| Djibouti | 1 | 0 |
| Egypt | 63 | 23 |
| Indonesia | 141 | 115 |
| Iraq | 3 | 2 |
| Laos PDR | 2 | 2 |
| Myanmar | 1 | 0 |
| Nigeria | 1 | 1 |
| Pakistan | 3 | 1 |
| Thailand | 25 | 17 |
| Turkey | 12 | 4 |
| Vietnam | 110 | 55 |
| Total | 417 | 257 |
Treatment
As for all influenza strains, there are several antiviral drugs available to reduce the duration and severity of flu caused by the H5N1 virus. A popular drug for treating this strain of flu is the neuraminidase inhibitor Oseltamivir (marketed as Tamiflu). This drug is potentially efficacious because it not only targets proteins that are common to all influenza A strains but also targets proteins especially important in the H5N1 genetic structure (7).
However, due to rapid mutation of the H5N1virus (8) and poor agricultural practices, there already are viral strains that are immune to some drugs. Drugs targeting M2 viral coat protein (amantadine or rimantadine) are generally cheaper than neuraminidase inhibitors like Tamiflu, although less effective. They were therefore originally chosen by the World Health Organization (WHO) to combat H5N1. When the drugs were found to be ineffective, it was discovered that poultry farmers in South China commonly administered these drugs to poultry, thus allowing H5N1 to evolve to become immune to this particular class of drugs. As a result in 2005, pharmaceutical company Hoffman-La Roche donated 3 million doses of Tamiflu to the WHO to prevent a pandemic (7,8).
Pandemic Prevention
Currently, the WHO conducts extensive monitoring for the H5N1 virus. In 2005, the WHO released a document recommending a number of “strategic actions” to prevent a pandemic from H5N1, and should it occur, to contain it as best as possible (9). One of the pre-pandemic objectives is to “reduce the opportunities for human infection” (9) through vaccinating all at-risk workers (9). This would significantly reduce the most likely transmission route by which the H5N1 virus could become a virus capable of human to human transmission (8). Over time, as more people are infected, the chances of a genetic exchange between human flu virus and avian flu virus will increase, thus allowing the avian flu virus more opportunities to create a transmissible human flu virus. Should a pandemic virus emerge, the WHO has plans to take steps to “contain or delay spread at the source. As part of this goal of slowing evolution of the virus, approximately 150 million birds either infected or at extreme risk of infection have been “destroyed” since 2003 (8).
The most pressing concern is that the extremely virulent H5N1 avian virus could acquire one of the hemagglutinins reactive with human cells and thus be carried by humans. The most straightforward way for this to occur would be for a human flu virus and the H5N1 avian flu virus to exist at the same time in a pig. Pigs have sugar chains necessary for both the human virus and the avian virus to flourish and whenever both viruses inhabit a pig at the same time there is a chance that the viruses will trade genes resulting in a human form of the H5N1 avian flu (10).
Several countries are in the process of developing human vaccines against the H5N1 virus, which would significantly hinder disease progress. However, typical seasonal flu can change significantly from year to year rendering previous vaccines ineffective, and the H5N1 virus mutates at an even higher rate. Thus by the time a vaccine is implemented the virus may have changed sufficiently to render the vaccine less than fully effective (8). This makes H5N1 especially dangerous as there is currently no technology capable of keeping up with its natural evolution.
According to the WHO, the risk of pandemic H5N1 is serious (8). Two of the three criteria for a pandemic as described by the WHO have been met: H5N1 is a new virus subtype and it causes serious human illness. The last criterion, that it can spread easily and sustainably from person-to-person, has not yet occurred. If it does, another pandemic, even more deadly than the “Spanish flu” pandemic, could occur.
References
1. “Evolution of H5N1 Avian Influenza Viruses in Asia.” http://www.cdc.gov/ncidod/EID/vol11no10/05-0644.htm
2. “Cumulative Number of Confirmed Human Cases of Avian Influenza A/(H5N1) Reported to WHO,” http://www.who.int/csr/disease/avian_influenza/country/cases_table_2009_04_08/en/index.html
3. Taubenberger, JK., and D.M. Morens “1918 Influenza: The Mother of All Pandemics.” The Center for Disease Control. 2005. Emerging Infectious Diseases. 8 April 2009. <http://www.cdc.gov/ncidod/eid/vol12no01/05-0979.htm#cit>.
4. “Key Facts About Avian Influenza (Bird Flu) and Avian Influenza A (H5N1) Virus,” http://www.cdc.gov/flu/avian/gen-info/facts.htm 7
5. “Flu Pandemic Morbidity / Mortality,” Globalsecurity.org. http://www.globalsecurity.org/security/ops/hsc-scen-3_flu-pandemic-deaths.htm
6. “H5N1 avian influenza: Timeline of major events,” World Health Organization, http://www.who.int/csr/disease/avian_influenza/Timeline_09_03_23.pdf
7. “Avian Influenza Mutation, H5N1,” http://www.avianinfluenza.org/mutated-avian-influenza-virus-h5n1.php
8. “Avian Influenza Frequently Asked Questions,” http://www.who.int/csr/disease/avian_influenza/avian_faqs/en/index.html
9. “Responding to the Avian Influenza Pandemic Threat: Recommended Strategic Actions.” World Health Organizations, http://www.who.int/csr/resources/publications/influenza/WHO_CDS_CSR_GIP_05_8-EN.pdf
10. Hatta, M., P. Gao, P. Halfmann and Y. Kawaoka (2001) “Molecular Basis for High Virulence of Hong Kong H5N1 Influenza A Viruses,” Science 293:1840-1842