Allochromatium vinosum
Classification
Domain (Bacteria); Phylum (Proteobacteria); Class (Gammaproteobacteria); Order (Chromatiales); family (Chromatiaceae); Genus (Allochromatium)
Use NCBI link to find]
Species

|
NCBI: Taxonomy |
Allochromatium vinosum
Description and Significance
“Allochromatium vinosum” is a gram negative, halotolerant, rod shaped, sulfide and thiosulfate oxidizing, anoxygenic purple sulfur bacteria that is generally 2.5-6.0 μm in length [1]. This organism’s habitat mainly consists of ponds, lakes, other stagnant bodies of fresh water, brackish waters, sewage lagoons, salt marshes, estuaries, and various marine habitats where hydrogen sulfide is exposed to light [3]. This metabolism relates to its environment in that it plays an important role in the reoxidation of sulfide from those bacteria, such as sulfate reducing bacteria, that like the deeper anoxic depths, along with being considered a primary producer of fixed carbon [2] . The previously mentioned oxidations take place under photolithoautotrophic conditions, and this motile organism also has the ability to undergo photoorganoheterotrophic growth [1] . The major ecological importance of this organism lies in its ability to removed sulfide and recycle elemental sulfur from environments deemed ‘sulfide-polluted’ with its biocatalytic properties, along with contributing to the continuation of the global sulfur cycle [4] .
Genome Structure
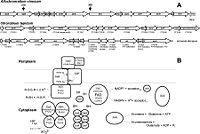
This organism has 3526903 chromosomes belonging to its circular chromosome. An interesting feature of this organism's gene sequence is that it contains carboxy-terminal DsrC cysteine residues which are important in the binding and transfer of sulfur-substrate, along with the electron transfer function involving thiol-disulfate interchanges [5] . Another interesting component of this organism’s genome is the dsr cluster; this cluster is comprised of 15 genes, “dsrABEFHCMKLJOPNRS”, which portions code for sulfite reductase and its various derivatives involved in oxidation of stored sulfur globules [11] .
Number of nucleotides: 3669074; Number of protein genes: 3220; Number of RNA genes: 64.
Based on the genome sequencing, it seems that there are 2 plasmids present in this specimen called pALVIN01 and pALVIN02, accounting for 102242 and 39929, respectively. The DNA base composition consisted of 61.3-66.3% mol G+C in reported strains. Also, this organism’s phylogenetic relation was based on its 16s rRNA gene groupings, specifically for pufL and pufM genes [6] .
Cell Structure, Metabolism and Life Cycle
This organism’s metabolism allows it to grow as a facultative photoautotroph as well as growing anaerobically in light with HS-, SO32-, S2O32-, and H2. “Allochromatium vinosum” has the ability to thrive in an aerobic chemotrophic manner without light with S2O32- and CO2 with low concentrations of oxygen. The process of sulfur oxidation occurs in the following fashion:
1. Oxidation of HS or S2O32- to elemental sulfur which is then put in the cell
2. Oxidation of HS- or elemental sulfur to SO32-
3. Oxidation of aforementioned molecule to resulting product of SO42- [7] .
In addition to the steps above, utilizing sulfite oxidoreductase, it is said that the organism has the ability to oxidize reduced sulfur to sulfate by APS (adenosine 5’-phosphosulfate) pathway [9] . This alternate pathway is important in that the organism has an additional pathway contributing to growth and enhanced electron flow and overall supply of reducing power [10] .
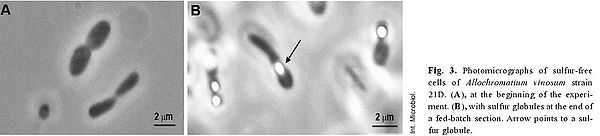
The above example of the organism’s metabolism interplays significantly with its cell structure and the sequential processes involved. This starts off by the factor of elemental sulfur being taken up by cell, forming globules of sulfur in periplasm as obligate intermediate preceding its oxidation to the final product of sulfate [8] . Somewhat in relation to cell structure, the dsr gene cluster, mentioned above, is quite important to the organism’s sulfur oxidation capabilities; this cluster is comprised of 15 genes, “dsrABEFHCMKLJOPNRS”, which portions code for sulfite reductase and its various derivatives involved in oxidation of stored sulfur globules [11] .
Ecology and Pathogenesis
As mentioned above, the varying habitats to which this organism belongs include freshwater, brackish water, and marine habitats [3] .
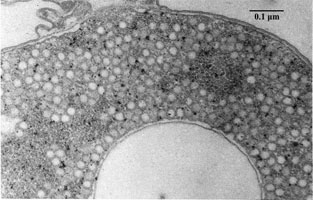
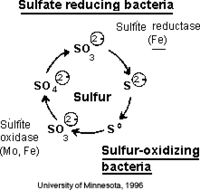
The biogeochemical significance of this organism is its role in global sulfur cycling. The importance lies in the fact the this organism has the ability to uptake relatively insoluble elemental sulfur; contributions to the cycle can be seen in above metabolism reactions [12] . In terms of contributions to the environment, phototrophic purple sulfur bacteria interplay with green sulfur bacteria and other organisms, facilitating the essential sulfur cycle as seen in Figure 4 [12] . A visual representation of sulfur globule formation is seen in Figure 6, below.
This organism has no reported data in regards to pathogenesis.
References
[2] Dahl, C. “’’Allochromatium vinosum’’ DSM 180-Home’’”. “DOE Joing Genome Project”. 2009.
[7] Paul, E. “’’Soil microbiology, ecology, and biochemistry’’”. 3rd edition. P. 410-412.
Author
Page authored by Anthony Rademacher and Polly Saavedra, students of Prof. Jay Lennon at Michigan State University.
