The Role of Helicobacter pylori in Intestinal Disease
Introduction
By Makarios McCune
Helicobacter pylori was first isolated by both Warren and Marshal in 1983[1]. The bacteria Helicobacter pylori is a rod shaped, gram negative bacterium equipped with a flagella for chemotaxis [2]. The bacterium is estimated to have infected 4.4 billion individuals worldwide, and is infamous for its association with numerous intestinal diseases and cancer. The bacterium typically infects the stomach and duodenum of the small intestine, and has been known to cause peptic ulcers and gastritis. If left untreated, the inflammation H. pylori can lead to stomach cancer and other serious health complications [3]. H. pylori is able to entrench itself into the gastric mucus of the stomach by producing urease to alter its morphology. Once situated, the bacterium can avoid detection from the immune system and propagate anti-inflammatory responses. However, Toll-like receptors (TLRs) are able to bind and recognize the bacterium and initiate inflammation. Unfortunately, the chronic inflammation produced by infection can lead to the upregulation of specific proteins and growth factors that increase the ability of cancer associated fibroblasts to metastasize throughout the body [3]. Infection is typically identified with either a stool antigen test, or a urea breath test. Endoscopies are also used to uncover infection, usually while also scoping out patients with chronic abdominal pain. Treatment for H. pylori infection involves an eradication program, requiring daily intake of various drugs and antibiotics to neutralize the infection. While generally successful, the development of antibiotic resistant strains has reduced the efficacy of some antibiotics - especially clarithromycin and nitroimidazole compounds. The growth of antibiotic resistant strains is due to the excretion of antibiotics into the environment, as well excessive use of antibiotics and negligence regarding following treatment plans. Eradication therapy can also leave lasting effects on the gut microbiome of the patient, as proton pump inhibitors and antibiotics can alter the pH of the environment and eliminate other species native to the stomach. Overtime, this dysbiosis can generate metabolic disorders and other digestive complications. Current solutions to this issue revolve around testing antimicrobial susceptibilities, and redesigning multi-drug eradication therapies.
Morphology
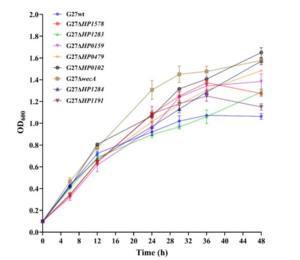
H. pylori is a gram negative bacterium that contains an outer membrane lined with lipopolysaccharide (LPS) in the outer leaflet [4]. This coating of anchored LPS helps the bacterium maintain structural integrity, membrane permeability, and avoid detection from Toll-like receptor 4 (TLR4) in the innate immune system [5]. Despite LPS’s importance to the bacterium’s survival, mutation to the formational glycosyltransferase proteins does not significantly impact bacterial fitness [4]. Truncation to multiple glycosyltransferase genes found did not impact the bacterial growth when measured under a OD600 assay [4]. Mutation to the LPS did however, alter the general shape of the bacteria. Deletion of the O antigen and Lewis antigen from the LPS structure changed the bacterial samples from a standard rod-shape to a cocoid shape [4]. Furthermore, removal of the O antigen also lead to a decrease in the minimum inhibitory concentration of polymxyin B needed to halt the growth of H. pylori, demonstrating the importance of LPS structural integrity for antimicrobial defense [4]. This trend was also seen with antibiotics such as clarithromycin, tetracycline, and amoxicillin, rifampicin, metronidazole, and levofloxcin also demonstrating low MICs against the truncated mutants [4]. This indicates that susceptibilities in the outer membrane can serve as prime targets for antibiotic treatment against vulnerable strains.
Besides mutation, H. pylori has also been known to change its morphology in response to its environment. For example, the bacterium has been observed switching from its standard rod shape to coccoid during bouts of nutrient starvation, temperature change, and in response to proton pump inhibitors [6]. These coccoid forms can coexist with the rod forms in the stomach and continue to produce urease and gastritis [6] [7]. The coccoid form has also been linked to a higher likelihood of cancer formation [8]. H. pylori can also adopt a filamentous form in hyperosmotic environments, and has been theorized to help the bacterium better adhere to the mucosal surfaces [6] [9]. The ability to switch to these differing morphs allows the bacterium to better adapt to environmental conditions and prolong its presence in the gut.
Infection and Immune Response
H. pylori infects around 50% or the worlds total population, with there being an estimated 4.4 billion cases around the globe [5] [2] [10]. Infection usually occurs orally, from consuming infected food and water sources [5]. Once inside the body, H. pylori migrates to the digestive system, and is prominently found in the gastric mucus [5]. Typically, the acidic environment of the stomach is able to neutralize bacteria and pathogens that manage to enter the body. H. pylori circumvents this issue through urease production and chemotaxis to infiltrate the gastric mucus and escape the harsh environment of the stomach lumen [5]. The urease catalyzes the production of of ammonium ions that interact with the gastric mucus and raises the pH of the system to neutral levels [5]. This alters the viscoelasticity of the structure and allows H. pylori to pass through rather than become trapped and degraded by antimicrobial agents [5].
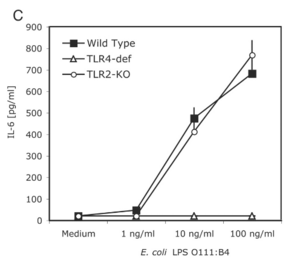
H. pylori employs a variety of unique strategies to thwart the mechanisms of the body’s immune system. One such strategy is the use of specific protein such as catalse and superoxide dismutase, which are responsible for counteracting the effects of reactive oxygen species deployed by the immune system [5]. The production of arginase is also used to neutralize nitric oxide production in macrophages and neutrophils that may target the bacterium [5]. Additionally, H. pylori is able to use to the gastric mucus produced by epithelial cells to hide its pathogen-associated molecular patterns (PAMPs) and from detection by pattern recognition receptors (PRRs) on immune cells [5]. The lipopolysaccharide (LPS) that lines the outer membrane of H. pylori also assists in defense from the immune system. The LPS on H. pylori is structured to lack phosphate groups on the 1’ and 4’ sections oft the lipid backbone, causing it to be less negatively charged [5]. This small change is enough to prevent binding and recognition of the LPS by Toll-like receptors (TLRs) [5]. However, this mechanism is not completely fool-proof, with a study finding that TLR4 could recognize the and bind the to LPS of the bacterium [10]. On the other hand, TLR2 could not detect plain LPS, but could recognize the entire intact bacterium, as well as other species of Helicobacter [10]. In this study, mice macrophages deficient in TLR4 could still mount an immune response and secrete IL-6, whereas TLR2 knockout cells could not [10]. This could be because TLR2 specializes in binding lipoproteins [5], making it unable to trigger a cytokine response to solely LPS.
TLR4 T and The flagellin protein belonging to the flagella of H. pylori are also unable to bind to TLR5, owing to a mutation in the N-terminal domain of the protein [5]. Additionally, H. pylori is able to prevent the induction of pro-inflammatory resposnes in the immune system. The fucosylated ligands of H. pylori are able to foil the C-type lectin receptor DC-SIGN - disrupting the protein complexes and halting downstream signalling for the pro-inflammatory response [5]. If uptaken by dendritic cells, H. pylori DNA can activate TLR9, which generates an anti-inflammatory response to prolong infection [5].
H. pylori can also disrupt action by the adaptive immune system by modulating responses from T cells. The virulence factor VacA, can disorder the interaction within the T cell receptor-IL2 pathway, which hampers T cell proliferation [5]. VacA has also been found to block nuclear translocation of the the transcription factor NFAT, which is required to activate many T cell related genes [5]. Moreover, VacA can force dendritic cells to upregulate the production of T regulator cells and IL-10 [5]). This causes T regulator cells to accumulate in large numbers in the gastric mucus, which lowers overall production of T helper cells and makes it far more difficult for them to eliminate invading H. pylori [5].
Despite its innate ability to avoid detection by the immune system, H. pylori is still vulnerable to being bound by NOD1 and NOD2 receptors [11]. The NOD1 receptor is able to bind and recognize peptidoglycan from H. pylori. Once bound, NOD1 signals for the transcription factor NF-κB, which subsequently causes gastric epithelial cells to secrete β-defensin 2 peptide which terminates H. pylori [12].
Cancer Development

H. pylori is strongly associated with the development of intestinal cancers, and is classified as a group 1 carcinogen by the World Health Organization (WHO) [13]. It has been estimated that those infected with H. pylori are three times more likely to be diagnosed with gastric cancer [13]. Gastric cancer itself is ranks fourth for cancer deaths worldwide [3]. H. pylori causes cancer by stimulating a pro-inflammatory response in the body. The continual persistence of the bacteria leads to chronic inflammation, which then leads to gastric cancer if left untreated. A study found that NFκB activated by infection activates an calcium channel protein called PIEZO1 [3]. Once activated, PIEZO1 can interact with other proteins like YAP1, which was found to increase metastasis in mouse samples [3]. Mice with knockout PIEZO1 saw a decrease in YAP1 expression, with YAP1 knockouts having nullified cancer cell migration [3]. Additionally, upregulation of PIEZO1 and YAP1 proteins was found to correspond to an increase presence of connective tissue growth factor (CTGF), which together with YAP1 and PIEZO1, increased the invasiveness of α-smooth muscle actin cancer associated fibroblasts [3]. These fibroblasts can then generate extracellular matrix that can lead to stiffening of cellular tissue by creating tension within tumors [14]. Stiffening alters the function of affected cells, and encourages them to migrate, also increasing metastasis of the cancer [3]. Moreover, PIEZO1 and the presence of the cancerous fibroblasts were both positively correlated to infection by H. pylori [3]. This demonstrates that H. pylori infection, the prevalence of calcium channel proteins, hardening of tumor cells, and metastasis of gastric cancer are all intertwined.
Treatment and Complications
The primary and most effective method for handling H. pylori infection is a combination of various identification techniques coupled with eradication treatment. For identification, both the stool antigen test and the urea breath test are typically the most effective and reliable for detecting H. pylori presence, and are recommended by American national standards. [13]. The stool antigen tests use monoclonal antibodies to detect antigens belonging to H. pylori in the patient’s stool samples [13]. Meanwhile, the urea breath test requires a patient to consume a pill or liquid containing urea, which is broken down into carbon dioxide by H. pylori. Excess carbon dioxide expelled by the patient is measured to determine the possibility of infection. Endoscopy can also be utilized to confirm H. pylori presence. However, it is suggested that endoscopies should primarily be reserved for determining the pathophysiology of patients with persistent upper abdominal pain, and not solely be used to detect H. pylori due to the invasive nature of the procedure [13].
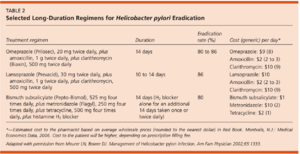
Once identified, H. pylori infection is typically cleared through an H. pylori eradication program. The treatment involves taking a mix of proton pump inhibitors, bismuth salts, and antibiotics for a duration of approximately 10-14 days [13]. However, treatment can vary greatly, with some therapies lasting for upwards of six weeks [13]. This combination drug therapy has proven to be potent - with patients reporting an eradication of upwards of 95% [13]. The number and type of drugs differs with regard to region, with different parts of the world having different standards for H. pylori eradication. North America, Europe, and China prescribe a quadruple drug therapy consisting of two or three antibiotics coupled with a proton pump inhibitor and sometimes bismuth. South Korea and Japan both still rely on the usage of just three drugs, with Japan basing treatment around the gastric suppressant drug vonoprazan [2]. Despite using less drugs, the vonoprazan regimen has proven to be exceedingly effective, with an eradication rate of 89-93%, and a greater than 80% success rate against clarithromycin resistant strains [2]. The antibiotics tetracycline and metronidazole were historically used for treatment, with amoxicillin, tinidazole, and clarithromycin now being utilized [2].
The largest obstacle for treatment currently is the development of antibiotic resistant strains. Antibiotic resistant H. pylori have been a growing concern, especially in the developed world [13]. WHO lists clarithromycin resistant H. pylori as a “high-priority species,” which is in the same category as methicillin resistant Staphylococcus aureus (MRSA) [2]. Approximately 10-30% of eradication treatments fail to eliminate H. pylori infection [1]. Specifically, nitroimidazole resistance is reported to be 10-56% in western nations, and 80-90% in developing nations [1]. Strains resistant to clarithromycin and nitroimidazole do significantly reduce the eradication percentage of treatment [1]. An extensive review of treatment literature found that H. pylori resistant to clarithromycin and nitroimidazole compounds have cure rates as low as 0% when treated with certain drug combinations [1]. The study also concluded that the cure rates for treating patients with nitroimidazole resistant H. pylori have decreased overall by 50% [1]. This problem of antibiotic resistance is exacerbated by the lack of a simplified method for testing H. pylori resistance, and the fact that multiple resistant strains can coexist within the human body at once [1]. Antimicrobial susceptibility testing is a common procedure that has proven helpful for determining the most effective treatment option for patients, as well as reducing unnecessary antibiotic usage [2]. Unfortunately, this method does require invasive endoscopy for sample acquisition, and is therefore not a standard medical procedure. Culturing and analyzing H. pylori also requires specialized laboratory equipment, which may be unavailable or costly to implement [2]. While a clear method for combatting resistant strains is still not fully fleshed out, a study did find treatment with rifabutin was 38% in clearing resistant cases [13]. The use of quadruple drug therapies has also been suggested as a possible solution and has been reported to have some success - although this was tested in a small patient testing pool [13] [1]. Future research and continued collaboration will be crucial for identifying strains of antibiotic-resistant H. pylori, and employing designing new drugs and therapies for eradication.
Besides antibiotic-resistance, gut microbiome dysbiosis is another crucial issue tied to H. pylori treatment. Dysbiosis is an imbalance to the local microbiome caused by the introduction or elimination of microorganisms. Because eradication therapy employs numerous antibiotics and gastric acid suppressants, the gut microbiota are directly impacted [2]. Studies recording the effects of eradication treatments found the alpha diversity indexes of patients to be lower 2 weeks following treatment [2]. Furthermore, the microbiome of groups receiving a quadruple drug treatment failed to return to baseline values even after a full year of recovery [2]. Even treatment with vonoprazan saw significant alterations to patients’ microbiomes last for two months before returning to standard levels [2]. It is clear that the attempt to treat an ongoing H. pylori infection comes with the added risk of reshaping the local gut microbiome and possibly causing unexpected metabolic and intestinal issues.
Additionally, H. pylori infection was found to conflict with other popular treatments, such as the usage of nonsteroidal anti-inflammatory drugs (NSAIDS) to treat dyspepsia and inflammation in the gut. Taking NSAIDS with an active H. pylori infection were found to increase the risk for both ulcer bleeding and peptic ulcer disease in infected patients [13]. Because of this, it is advised that patients diagnosed with dyspepsia and H. pylori undergo eradication therapy before beginning chronic NSAIDS therapy [13]. Other drug-related complexities require specific planning with medical professionals.
Conclusion
In conclusion, H. pylori is a bacterium with the potential to cause peptic ulcers, gastric cancers, and various intestinal diseases. The most common treatment option for infection is eradication therapy that requires patients to take multiple drugs and antibiotics over a 10-14 day period. Current eradication rates for this treatment range from 80-90%, however the continual emergence of antibiotic resistant strains of H. pylori threaten to neutralize this effectiveness as the prevalence of these strains begins to proliferate. In order to improve treatment options, limit the intestinal dysbiosis, and combat the rise of antibiotic resistance, new drug therapies need to be tested, and affordable and efficient methods of strain resistance need to be researched.
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 Houben, Beek, D.V.D., Hensen, Craen, A.J.M.D., Rauws, Tytgat, 1999. A systematic review of Helicobacter pylori eradication therapy—the impact of antimicrobial resistance on eradication rates. Aliment Pharmacol Ther 13, 1047–1055. https://doi.org/10.1046/j.1365-2036.1999.00555.x
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 Suzuki, S., Kusano, C., Horii, T., Ichijima, R., Ikehara, H., 2022. The Ideal Helicobacter pylori Treatment for the Present and the Future. Digestion 103, 62–68. https://doi.org/10.1159/000519413
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 3.7 3.8 Chen, B., Liu, X., Yu, P., Xie, F., Kwan, J.S.H., Chan, W.N., Fang, C., Zhang, J., Cheung, A.H.K., Chow, C., Leung, G.W.M., Leung, K.T., Shi, S., Zhang, B., Wang, S., Xu, D., Fu, K., Wong, C.C., Wu, W.K.K., Chan, M.W.Y., Tang, P.M.K., Tsang, C.M., Lo, K.W., Tse, G.M.K., Yu, J., To, K.F., Kang, W., 2023. H. pylori ‐induced NF‐κB‐PIEZO1‐YAP1‐CTGF axis drives gastric cancer progression and cancer‐associated fibroblast‐mediated tumour microenvironment remodelling. Clinical & Translational Med 13, e1481. https://doi.org/10.1002/ctm2.1481
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 Tang, X., Yang, T., Shen, Y., Song, X., Benghezal, M., Marshall, B.J., Tang, H., Li, H., 2023. Roles of Lipopolysaccharide Glycosyltransferases in Maintenance of Helicobacter pylori Morphology, Cell Wall Permeability, and Antimicrobial Susceptibilities. IJMS 24, 11381. https://doi.org/10.3390/ijms241411381
- ↑ 5.00 5.01 5.02 5.03 5.04 5.05 5.06 5.07 5.08 5.09 5.10 5.11 5.12 5.13 5.14 5.15 5.16 5.17 5.18 5.19 Salama, N.R., Hartung, M.L., Müller, A., 2013. Life in the human stomach: persistence strategies of the bacterial pathogen Helicobacter pylori. Nat Rev Microbiol 11, 385–399. https://doi.org/10.1038/nrmicro3016
- ↑ 6.0 6.1 6.2 Krzyżek, P., Gościniak, G., 2018. Morphology of Helicobacter pylori as a result of peptidoglycan and cytoskeleton rearrangements. pg 13, 182–195. https://doi.org/10.5114/pg.2018.78284
- ↑ She, F.-F., 2003. Virulence of water-induced coccoid Helicobacter pylori and its experimental infection in mice. WJG 9, 516. https://doi.org/10.3748/wjg.v9.i3.516
- ↑ Loke, M.F., Ng, C.G., Vilashni, Y., Lim, J., Ho, B., 2016. Understanding the dimorphic lifestyles of human gastric pathogen Helicobacter pylori using the SWATH-based proteomics approach. Sci Rep 6, 26784. https://doi.org/10.1038/srep26784
- ↑ Yang, D.C., Blair, K.M., Salama, N.R., 2016. Staying in Shape: the Impact of Cell Shape on Bacterial Survival in Diverse Environments. Microbiol Mol Biol Rev 80, 187–203. https://doi.org/10.1128/MMBR.00031-15
- ↑ 10.0 10.1 10.2 10.3 Mandell, L., Moran, A.P., Cocchiarella, A., Houghton, J., Taylor, N., Fox, J.G., Wang, T.C., Kurt-Jones, E.A., 2004. Intact Gram-Negative Helicobacter pylori , Helicobacter felis , and Helicobacter hepaticus Bacteria Activate Innate Immunity via Toll-Like Receptor 2 but Not Toll-Like Receptor 4. Infect Immun 72, 6446–6454. https://doi.org/10.1128/IAI.72.11.6446-6454.2004
- ↑ Kaparakis, M., Turnbull, L., Carneiro, L., Firth, S., Coleman, H.A., Parkington, H.C., Le Bourhis, L., Karrar, A., Viala, J., Mak, J., Hutton, M.L., Davies, J.K., Crack, P.J., Hertzog, P.J., Philpott, D.J., Girardin, S.E., Whitchurch, C.B., Ferrero, R.L., 2010. Bacterial membrane vesicles deliver peptidoglycan to NOD1 in epithelial cells. Cellular Microbiology 12, 372–385. https://doi.org/10.1111/j.1462-5822.2009.01404.x
- ↑ Grubman, A., Kaparakis, M., Viala, J., Allison, C., Badea, L., Karrar, A., Boneca, I.G., Le Bourhis, L., Reeve, S., Smith, I.A., Hartland, E.L., Philpott, D.J., Ferrero, R.L., 2010. The innate immune molecule, NOD1, regulates direct killing of Helicobacter pylori by antimicrobial peptides. Cellular Microbiology 12, 626–639.
- ↑ 13.00 13.01 13.02 13.03 13.04 13.05 13.06 13.07 13.08 13.09 13.10 13.11 13.12 Ables, A.Z., Simon, I., Melton, E.R., 2007. Update on Helicobacter pylori treatment. Am Fam Physician 75, 351–358.
- ↑ Benn, M.C., Pot, S.A., Moeller, J., Yamashita, T., Fonta, C.M., Orend, G., Kollmannsberger, P., Vogel, V., 2023. How the mechanobiology orchestrates the iterative and reciprocal ECM-cell cross-talk that drives microtissue growth. Sci. Adv. 9, eadd9275. https://doi.org/10.1126/sciadv.add9275
Authored for BIOL 238 Microbiology, taught by Joan Slonczewski,at Kenyon College,2024
