Coronavirus
A Viral Biorealm page on the genus Coronavirus
Baltimore Classification
Higher order taxa
Virus; ssRNA positive-strand viruses, no DNA stage; Nidovirales; Coronaviridae; Coronavirus
Species
Human coronavirus, SARS coronavirus, Bovine coronavirus, Canine coronavirus (examples)
Description and Significance
Severe Acute Respiratory Syndrome (SARS) is a strain of coronavirus in humans that is known for causing super-spreader events. These events occur when an individual infects others directly and indirectly, such as friends, family, and even hospital staff. The most significant SARS super-spreader event took place in a hotel in 2002, causing the hotel guests to unknowingly become global carriers of the virus. The outbreak of the SARS-CoV from 2002-2003 resulted in 774 deaths as well as staggering economic losses within the span of six months [3]. The virus had a global economic impact with the Asian Development Bank recording losses of 59 billion USD [3].
A few years later, the first case of Middle Eastern Respiratory Syndrome (MERS-CoV) was found and documented in Saudi Arabia. The MERS-CoV virus was sequenced and identified as related to SARS-CoV [4]. Due to the known pandemic potential of SARS in addition to the annual religious pilgrimage to Mecca, MERS triggered global health security concerns. However, epidemiological reports showed that MERS-CoV is less contagious and has a lower pandemic potential than SARS [4]. Despite the lower pandemic potential, MERS-CoV, like SARS-CoV, also had a global impact. Travel restrictions applied to all travelers in the infected area, thus interfering with the normal traffic to and through the region [4]. Due to the cascade of global public health concerns and economic losses caused by strains of the Coronavirus, contemporary research strives to discover preventative drugs and to educate the public on the effects of SARS and MERS [3,4].
History and Classification
The Coronaviridae family has been long studied since the isolation of the prototype murine coronavirus strain JHM in the late 1940’s and are known to affect both animal and human species [5]. The porcine transmissible gastroenteritis virus (TGEV), bovine coronavirus (BCoV), and avian infectious bronchitis viruses (IBV) are important for veterinary studies, and the murine coronavirus mouse hepatitis virus (MHV) is studied as a model mechanism for human diseases [5]. In the 1960’s, studies of human coronaviruses were found to cause common cold symptoms. However, in the Spring of 2003, a new strand of human coronavirus emerged, which was recognized worldwide as the SARS epidemic [5]. Due to the increased severity of the new human coronavirus strains, coronaviruses are now considered as emerging pathogens [5]. There are three genera of coronaviruses: group I includes animal pathogens such as TGEV; group II includes viruses of veterinary importance (ex. BCoV) and respiratory-related human coronaviruses; and group III includes avian coronaviruses [5].
There has been controversy as to whether the emerging pathogens such as SARS-CoV belongs in group II or in its own genera [5]. In order to classify and or differentiate SARS-CoV as a part of Group II or in its own genera entirely, sequence comparisons were performed between three well-defined enzymatic proteins from SARS-CoV and already well-characterized coronaviruses [6]. The comparisons showed that while the phylograms were similar in regards to the experimentally tested protein, that the SARS-CoV sequences diverged into a fourth distinct branch and were overall dissimilar from prior coronaviruses, therefore, not belonging under the group II genera [6].
Cell Structure
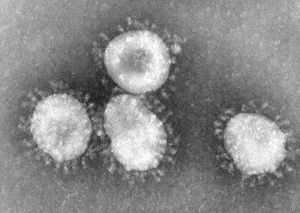
An identifiable trait of coronaviruses is the club-shaped spikes, resembling solar coronas, that project from the cell membrane. [2]. The nucleocapsid of coronaviruses has a helical shape, which is uncommon for positive-sense RNA viruses [2]. The virion shape is spherical, with an average size of 125 nm [2]. There are four main structural proteins: spike, membrane, envelope, and nucleocapsid proteins [2]. The membrane protein is the most common, and is thought to be responsible for the spherical shape of the virion [2]. Viruses that do not have the envelope protein expression are not lethal, though further research is needed to confirm this hypothesis [2]. The envelope protein can vary among different species of coronavirus, but its responsibilities include gathering and releasing the virus to allow it to infect a host cell.
Coronaviruses replicate in the host cell and then induce changes that can be identified by scientists [7]. For example, coronaviruses cause the endoplasmic reticulum of the host cell to convolute in specific areas. This mechanism allows the virus to be protected against antiviral mechanisms produced by the host’s immune system [7]. This, combined with the characteristic spikes of the virus, are important structures that allow the virus to continue surviving and causing harm to a host organism.
Genome Structure
Coronaviruses belong to the Nidovirales family, which is characterized by an enveloped, positive-sense RNA virus [2]. Compared to other viruses, coronaviruses have a relatively long genome of 30 kilobases [2]. Twenty of these kilobases encode non-structural proteins, and the remaining ten of these kilobases are responsible for structure and accessory functions. The genome for coronaviruses also includes a 5’ cap and 3’ poly-A tail, which allow the virus to act as an mRNA molecule to translate replicase polyproteins [2]. The function of accessory proteins is not entirely known, but one possible hypothesis is that they are used for viral pathogenesis [2].
Further research shows some coronaviruses lack exoribonuclease activity, which can limit their ability to infect a host cell [8]. Microorganisms without exoribonuclease activity are able to increase the rate of mutations compared to microorganisms with exoribonuclease activity [8]. Furthermore, coronaviruses lacking exoribonuclease activity are less able to invade host organisms [8]. Exoribonuclease expression in the genome is critical in order for the virus to replicate; therefore, the genes for expression of exoribonuclease may be a viable target for inhibition in future vaccinations for coronavirus [8].
Metabolic processes
Coronavirus mRNAs are generated through a complex method of discontinuous extension, which occurs during the synthesis of minus-strand subgenome-length templates [9]. The replicase-transcriptase proteins, along with other viral proteins, manifest into membrane-bound replication-transcription complexes (RTC) [9]. The RTCs are encoded in open-reading frame 1a (ORF1a) and ORF1b and carry the viral N protein. The implication that N protein was important for coronavirus virulence was observed by performing experiments with the absence of N protein, which resulted to little or no amplification of the replicon [9].
In addition, the SARS-CoV uses another mechanism to increase the viral fitness and metabolic processes. The envelope (E) gene of the SARS-CoV encodes a small multifunctional protein that possesses ion channel activity, which is an important mechanism for virus-host interaction [10]. They found that deletion of the E gene, mitigates the virus as a result of making the IC defective. To test their finding, they used two recombinant mice with adapted SARS-CoVs, each carrying an intact IC and the other with a mutation that rendered the IC inactive [10]. Those with the active IC resulted in death of mice; and those with mutant E gene lacking IC activity, generally resulted in recovery and survival of the mice [10]. However, it was noted that it was not the knocking down of the E protein that demonstrated significant effect to the virulence. Rather, it was the decreased major symptom of edema accumulation, the primary cause of death for acute respiratory distress syndrome and often caused by SARS-CoV, that displayed importance.
The genome replication of coronavirus is mediated through a full-length minus-strand copy of the genome that serves as a template for the production of progeny virus genomes. The transcription of coronavirus is a complex process involving the discontinuous synthesis of up to eight minus-strand RNAs of subgenomic size which contain sequences corresponding to the 5' and 3' ends of the genome and serve as templates for the synthesis of a 5'- and 3'- coterminal set of subgenomic mRNAs. According to the current model of coronavirus discontinuous extension, the synthesis of nascent minus-strand RNA initiates at the 3- end of the plus-strand genomic template and proceeds to TRS elements that are located at various sites within the virus genome. A proportion of the nascent minus-strand RNA is translocated to the 5' leader sequence of the genome, and subsequently a minus-strand copy of the leader is added at these sites. The TRS elements thus determine the fusion sites of leader and "body"-derived sequences of the subgenomic RNAs, and the number of TRS elements correlates with the number of subgenomic mRNAs produced by a particular coronavirus.
The proteins encoded by the replicase gene are critically involved in both replication and transcription. The coronavirus replicase proteins are synthesized as large polyproteins that are extensively processed by virus-encoded proteinases to produce a functional replicase or transcriptase complex. Suggestions have been made that this complex comprises so-called scaffolding proteins, together with proteins that have enzymatic functions, including RNA-dependent RNA polymerase, helicase, and uridylate-specific endoribonuiclease (NendoU), and putative exonuclease and 2'-O-ribose methyl transferase activities. The active replicase or transcriptase complex may contain other proteins of viral or cellular origin in addition.
Viral Ecology
Understanding the ecology of the coronavirus involves examining its relationships with other organisms (bats and camels) as well as the physical environments (respiratory and gastrointestinal tracts) where it thrives. Both hallmark strains of the coronavirus, SARS-CoV and MERS-CoV, originated zoonotically - in particular, bats and camels respectively. It is characteristic of RNA viruses to replicate in its reservoir – a long term host species – at a constant rate [11]. However, interspecies transmission triggers the virus to replicate at an exponential rate in its new host [11]. According to phylogenetic and dating studies, Vijaykrishna and Smith (2007) have shown that coronavirus undergo a constant population growth in bats, but when transmitted to other hosts, the virus displays an epidemic like increase in population, leading the researchers to identify bats as the reservoir for this virus [11].
As to the physical environments of coronaviruses, they have been found to reside more commonly in the respiratory and gastrointestinal tracts in their human hosts; therefore, coronaviruses have been linked to a range of mild to fatal upper and lower respiratory tract illnesses, and additionally play a mild role in acute gastroenteritis (AGE) [12]. Furthermore, most coronavirus cases were detected in the winter, a moderate amount of cases were detected in the spring and autumn, and no cases were detected in the summer, indicating a correlation between weather changes and the transmission of the coronavirus [12].
Viral Pathology
There are several theories regarding SARS transmission and how the infectious process manifests in the host body. After initial contact with the virus, injury to lungs is then followed by dissemination of the virus throughout the body [13]. The SARS virus is transmitted via respiratory droplets, direct contact with fomites, and aerosolized respiratory secretions [14]. Once inside the host body, the virus targets the epithelial cells, mainly in the respiratory tract, but not limited to the mucosal cells of the intestines, tubular epithelial cells of the kidneys, and neurons of the brain [13].
In a study performed in Hong Kong, the researchers found that the average incubation period of the disease was approximately 6.4 days, and the average time from onset of flu-like symptoms to hospital was 3-5 days [14]. Those that were over the age of 60 had a significantly higher fatality rate of 43% versus 6.8% in younger patients, indicating that survivability is strongly influenced by patient age [14]. The predominant pathological finding in autopsies was diffuse alveolar damage (DAD); however, it is also difficult to distinguish DAD as a result from SARS versus trauma, aspiration, oxygen toxicity, or other infectious microorganisms [13]. In further research, SARS has been observed to be not isolated as a respiratory viral infection, but also as systemic virus, involving damage to the spleen, liver, intestines, kidneys, heart, and adrenal gland [16].
Current Research
Current research on the coronavirus works continuously to combat MERS-CoV and SARS-CoV in humans. Specifically, ongoing research aims to discover a disease model that is able to express the disease severely and in a uniform manner [17]. By discovering the ideal disease model, researchers would then be able to develop preventative and therapeutic drugs to prevent a re-emergence of SARS or MERS. Research to prevent another global outbreak is of particular importance as there is currently no licensed preventive or therapeutic drug to use in a future outbreak of SARS or MERS [18]. Additionally, research is working toward understanding the role of domestic livestock and other species in disease transmission to comprehend the epidemiology of the coronavirus [18]. Understanding the role of domestic livestock in transmitting the disease also enables animal-owners to vaccinate their animals to prevent another SARS or MERS outbreak [18]. Researching preventative and therapeutic drugs as well as understanding the zoonotic transmission can be utilized to prevent future global outbreaks of SARS or MERS.
After the 2015 outbreak of MERS in Qatar, researchers investigated zoonotic sources for the transmission of MERS-CoV [18]. The prevalence of domestic livestock in the Middle East led researchers to investigate these animals specifically; livestock were also evaluated for their advantages and limitations as animal models for viral pathogenesis and transmission studies [18]. The viral RNA was only discovered in dromedaries and alpacas, which are suspected to be the main source of zoonotic transmission [18]. Phylogenetic analyses discovered that MERS-CoV was created in dromedary camels by gene recombination, which is suspected to have led to the 2015 outbreak in Qatar [18]. One method to administer a MERS-CoV vaccine effectively is to combine the vaccine with the camelpox virus vaccine for camel owners to give to their animals in the Middle East, Africa, and Asia [18]. A combination vaccine of rabies virus and MERS-CoV was also designed since the rabies vaccine is already administered to camels. These vaccines would be an effective preventative measure against the zoonotic transmission of MERS [18].
Model species are an effective method as an in vivo model to test the efficacy of vaccines and treatments before applying them to humans. Camels are natural hosts which makes them an appropriate model species; however, these animals are extremely costly and difficult to work with due to their size [18]. Macaques and marmosets can be utilized to develop the clinical disease similar to humans, but these species involve complex husbandry that increases costs [17].
Despite the increased cost of using nonhuman primates, the marmoset was tested as a model for MERS-CoV in humans. Falzarano et al. (2014) discovered that most animals developed a progressive severe pneumonia. Necropsies showed high viral loads and lesions in the lungs of all animals. RNA-seq confirmed the induction of immune and inflammatory pathways [17]. Falzarano et al. (2014) successfully investigated the ability of the marmoset to model MERS-CoV by confirming the presence of the viral disease symptomatically and through RNA-seq. The progression of the disease was analyzed and concluded that the marmoset is a severe, partially lethal disease model of MERS-CoV. Severe disease models enable researchers to discover treatments and combat the disease at a high standard [17]. The model can be utilized to test the efficiency of vaccines and treatments strategies that can be applied to humans [17]. Continuing research of MERS, and the closely related SARS, works to develop a complete list of potential host targets as well as develop preventative and therapeutic drugs to prevent a re-emergence of the disease [18].
References
1. “Home - Taxonomy - NCBI.” National Center for Biotechnology Information, U.S. National Library of Medicine, www.ncbi.nlm.nih.gov/taxonomy [Webpage]. (Assessed Nov. 1, 2017)
2. Fehr, A.R., and Perlman, S. 2015. Coronaviruses: An Overview of Their Replication and Pathogenesis. Methods in Molecular Biology. 1282: 1–23.
3. Baric, R.S. 2008. SARS-CoV: Lessons for global health. Virus Research. 133(1): 1-3.
4. Al-Tawfiq, J.A., Zumla, A., and Memish, Z.A. 2014. Travel implications of emerging coronaviruses: SARS and MERS-CoV. Travel Medicine and Infectious Disease. 12(5): 422-428.
5. Weiss, S.R., and Navas-Martin, S. 2005. Coronavirus Pathogenesis and the Emerging Pathogen Severe Acute Respiratory Syndrome Coronavirus. Microbiology and Molecular Biology Reviews. 69(4): 635-664.
6. Rota, P.A., Oberste, M.S., Monroe, S.S., Nix, W.A., Campagnoli, R., Icenogle, J.P., Peñaranda, S., Bankamp, B., Maher, K., Chen, M.H. et al. 2003. Characterization of a Novel Coronavirus Associated with Severe Acute Respiratory Syndrome. Science. 300(5624): 1394-1399.
7. Kikkert, M., Knoops, K., Koster, A., van den Worm, S., Zevenhoven-Dobbe, J., van der Meer, Y., Koster, A., Mommaas, A., and Snijder, E. 2008. SARS-Coronavirus Replication Is Supported by a Reticulovesicular Network of Modified Endoplasmic Reticulum. PLoS Pathogens. 6(9): e226.
8. Blanc, H., Denison, M., Smith, E., and Vignuzzi, M. 2013. Coronaviruses Lacking Exoribonuclease Activity Are Susceptible to Lethal Mutagenesis: Evidence for Proofreading and Potential Therapeutics. PLoS Pathogens. 9(8): e1003565.
9. Sawicki, S.G., Sawicki, D.L., and Siddell, S.G. 2007. A Contemporary View of Coronavirus Transcription. Journal of Virology. 81(1):20-29.
10. Nieto-Torres, J.L., DeDiego, M.L., Verdiá-Báguena, C., Jimenez-Guardeño, J.M., Regla-Nava, J.A., Fernandez-Delgado, R., Castaño-Rodriguez, C., Alcaraz, A., Torres, J., Aguilella, V.M., and Enjuanes, L. 2014. Severe Acute Respiratory Syndrome Coronavirus Envelope Protein Ion Channel Activity Promotes Virus Fitness and Pathogenesis. Denison MR, ed. PLoS Pathogens. 10(5).
11. Vijaykrishna, D., Smith, G.J.D., Zhang, J.X., Peiris, J.S.M., Chen, H., and Guan, Y. 2007. Evolutionary Insights into the Ecology of Coronaviruses. Journal of Virology. 81(8): 4012-4020.
12. Jevsnik, M., Steyer, A., Pokorn, M., Mrvič, T., Grosek, Š., Strie, F., Lusa, L., and Petrovec, M. 2016. "The Role of Human Coronaviruses in Children Hospitalized for Acute Bronchiolitis, Acute Gastroenteritis, and Febrile Seizures: A 2-Year Prospective Study." PLoS One. 11(5): e0155555.
13. Gu J., and Korteweg, C. 2007. Pathology and Pathogenesis of Severe Acute Respiratory Syndrome. The American Journal of Pathology. 170(4): 1136-1147.
14. Tsui, P.T., Kwok, M.L., Yuen, H., and Lai, S.T. 2003. Severe Acute Respiratory Syndrome: Clinical Outcome and Prognostic Correlates. Emerging Infectious Diseases. 2003 Sep; 9(9): 1064–1069.
15. Donnelly, C.A., Ghani, A.C., Leung, G.M., Hedley, A.J., Fraser, C., Riley, S., Abu-Raddad, L.J., Ho, L.M., Thach, T.Q., Chau, P., et al. Epidemiological determinants of spread of causal agent of severe acute respiratory syndrome in Hong Kong. The Lancet. 361(9371): 1761-1766.
16. Ding, Y., Wang, H., Shen, H., Li, Z., Geng, J., Han, H., Cai, J., Li, X., Kang, W., Weng, D., et al. 2003. The clinical pathology of severe acute respiratory syndrome (SARS): a report from China. Journal of Pathology. 200: 282–289.
17. Falzarano, D., de Wit, E., Feldmann, F., Rasmussen, A. L., Okumura, A., Peng, X., Thomas, M. J., Doremalen, N. V., Haddock, E., Nagy, E., et al. 2014. Infection with MERS-CoV causes lethal pneumonia in the common marmoset. PLoS pathogens, 10(8).
18. Vergara-Alert, J., Vidal, E., Bensaid, A., and Segalés, J. 2017. Searching for animal models and potential target species for emerging pathogens: Experience gained from Middle East respiratory syndrome (MERS) coronavirus. One Health. (3): 34-40.