Heavy Metal Toxicity and Resistance
Introduction
The widespread use of metals has prompted many microbiologists to examine the relationship between heavy metal toxicity and bacterial resistance. Heavy metals are defined as those which have a density of over 5g/cm3 (1), and thus include many of the transition and coinage metals. The reversion to heavy metal biocide use has been suggested as a possible solution to antibiotic resistance (2). On the other hand, bacteria that are resistant to certain heavy metals have been proposed to be economically useful in biomining and bioremediation (1). The constant race between heavy metal toxicity and bacterial resistance will likely give rise to many ingenious applications in the future, so it is important to understand the underlying mechanisms.
Heavy metal toxicity mechanisms
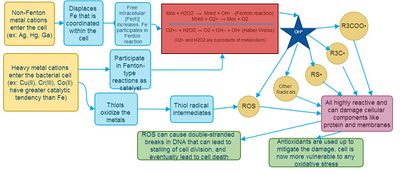
Heavy metals have been historically used as antimicrobial agents prior to the discovery of antibiotics, and have now been incorporated into coatings, surfaces, and internally placed devices (3). Recent evidence has shown that some metals can work synergistically with antibiotics (Cu with polycide) (4), are effective against multi-drug resistant bacteria, and can disrupt biofilms (5, 6). There is a lot of diversity in both metals and the bacteria they affect, but in general toxicity can be broken down into five categories (FIG 1).
Oxidative stress
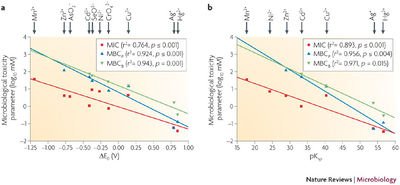
Within normal bacterial cells, iron is essential as a cofactor in enzymes, but it also available in cells to undergo the Fenton reaction to produce reactive oxygen species (ROS) that are damaging to all biological macromolecules (FIG 2) (7). In normal cells, only 20 μM of Fe2+ is available for Fenton reaction, because the rest are all engaged in coordination interactions with ligands (8). When extracellular Fe, Cu, or Ni is introduced into the cell, an increase in ROS and radicals is observed (2). Thiols within the cell also have a high affinity for reducing metals covalently, and can give rise to ROS (2). In order to buffer the redox state within the cell, antioxidants are depleted, and the cell is more vulnerable to ROS from normal metabolism byproducts. Therefore, the metal’s solubility and the reduction potential correlate with the toxicity within the cell (FIG 3) (9).
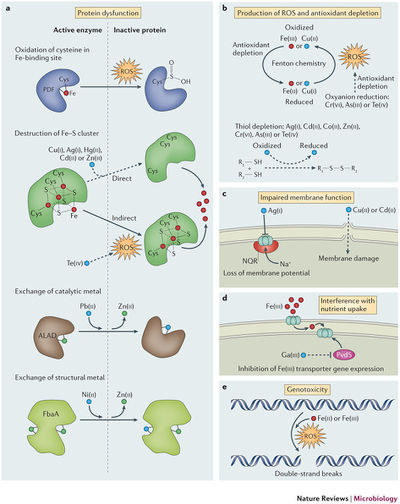
Protein activity interference
It has been noted that only a few amino acid alterations near the metal-binding region is required to disable function of an enzyme (10). The metal ions often oxidize residues and lead to a formation of carbonyl groups, and proteins with these marker groups are designated for intracellular degradation (11). In bacteria, a family of Fe-S dehydratases is extremely vulnerable to such ROS-mediated oxidation (12). Furthermore, thiol groups and disulfide groups that are important in substrate binding may also be oxidized by cations (3). There has also been evidence of metal cations displacing wild-type cofactors in enzymes using ionic or molecular mimicry - where the metal species is structurally similar but physiologically disruptive (2). An example of this would be gallium substitution of iron (FIG 4) in ribonucleotide reductase, the first enzyme in DNA replication. Ga has similar ionic radius to Fe, but no other oxidation state that is essential to enzyme activity. This substitution also inhibits transcription of pvds (Fe-responsive transcription regulator) in Pseudomonas aeruginosa, initiating a positive feedback cycle that is toxic to multidrug-resistant organisms and quiescent cells in the center of biofilms (5). This displacement is how Fenton-inactive metals like Ag can release coordinated, intracellular Fe into the cytoplasm and contribute to ROS formation (13).

Nutrient assimilation interference
Metal ions must first enter the cell in order to have an effect (1). They can be transported by fast, unspecific, constitutively-expressed transporters like MIT transporters, or specific, inducible ABC transporters and P-type transporters (1). Some cations can competitively inhibit the assimilation of essential ions by binding to these transporters (14). For example, Cr (IV) reduces sulphate uptake since chromate exhibits molecular mimicry, causing a decrease in intracellular S, and a deficiency in this essential bioelement (15).
Membrane impairment
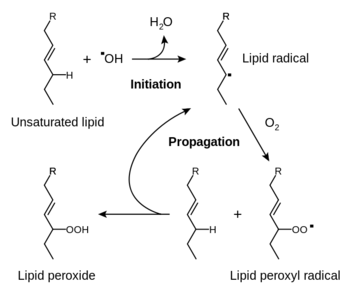
It has been hypothesized copper and cadmium can cause lipid peroxidation (FIG 4) by increasing mutations that increase the number of unsaturated bonds in fatty acids (16), affecting the integrity of the cytoplasmic membrane. Another method of metal toxicity is through dissipation of the chemiosmotic force or siphoning electrons from the ETC. Silver in Vibrio cholerae have been found to introduce “leakage” of protons across the membrane (17), while other metals are reduced by the quinone molecule and steal electrons away from respiration (3).
Genotoxicity
ROS that was previously created by the Fenton reaction can disrupt DNA replication and lead to cell death (8). While Cr(IV) is a very potent mutagen, lab situations have suggested many additional cations that cause DNA damage (2). Furthermore, copper has been correlated to the destruction of extracellular DNA following cell lysis, and this may inhibit postmortem horizontal gene transfer of resistance via transformation (18).
Resistance Mechanisms
Metal toxicity differs from antibiotics mostly because there are no components to modify (1), but there are still a couple of ways the cell can defend itself.
Efflux or sequestration
Cations can be pumped out of the cell with low ATP expense, or sequestered by chelates like thiol-containing compounds at approximately 16 ATP, an option that is only cost-effective at low metal concentrations (3).
Reduction to less toxic state
Some cations can be reduced to a less detrimental state if its reduction potential is within physiological conditions (-421 to 808mV), but sometimes the reduced state may be more toxic or insoluble (1). Extracellular electron transfer has also been hypothetically linked to nanowires, and is an area of active research (3).
Biofilm formation
Some bacteria like P. aeruginosa can form biofilms, where there is a heterogeneous population of cells with altered physiology (3). For example, those that are nearest to the substrate are usually anoxic and quiescent, and thus experience less ROS production, allowing tolerance to metal (19). Even dead cells can sequester metals from live cells of the biofilm (20). In a method similar to cell differentiation in eukaryotes, the biofilm can use quorum sensing to translate detoxifying enzymes or secrete an extracellular polymeric substance which biosorbs cations and impedes diffusion (21). The exact processes are still being examined, but with many phenotypic variants within a biofilm, the versatility and differentiation may give these bacteria an advantage against heavy metals.
Closing Word
Each metal ion has a different method of inflicting damage, while each tolerance mechanism has intrinsic advantages against particular cations. More research will help elucidate this arms race and help scientists harness them for use. Currently, heavy metals do seem like a plausible substitute for antibiotics, but its exact administration and mechanisms must be uncovered before clinical use.
References
1. Nies, D. H. 1999. Microbial heavy-metal resistance. Appl. Microbiol. Biotechnol. 51:730-50. doi: http://dx.doi.org.ezproxy.library.ubc.ca/10.1007/s002530051457.
2. Lemire, J. A., J. J. Harrison, and R. J. Turner. 2013. Antimicrobial activity of metals: mechanisms, molecular targets and applications. Nat Rev Micro. 11:371-384.
3. Harrison, J. J., H. Ceri, and R. J. Turner. 2007. Multimetal resistance and tolerance in microbial biofilms. Nat Rev Micro. 5:928-938.
4. Harrison, J. J., R. J. Turner, D. A. Joo, M. A. Stan, C. S. Chan, N. D. Allan, H. A. Vrionis, M. E. Olson, and H. Ceri. 2008. Copper and Quaternary Ammonium Cations Exert Synergistic Bactericidal and Antibiofilm Activity against Pseudomonas aeruginosa. Antimicrobial Agents and Chemotherapy. 52:2870-2881. doi: 10.1128/AAC.00203-08.
5. Kaneko, Y., M. Thoendel, O. Olakanmi, B. E. Britigan, and P. K. Singh. 2007. The transition metal gallium disrupts Pseudomonas aeruginosa iron metabolism and has antimicrobial and antibiofilm activity. J. Clin. Invest. 117:877-888. doi: 10.1172/JCI30783.
6. Harrison, J. J., H. Ceri, C. A. Stremick, and R. J. Turner. 2004. Biofilm susceptibility to metal toxicity. Environ. Microbiol. 6:1220-1227. doi: 10.1111/j.1462-2920.2004.00656.x.
7. Pomposiello, P. J., and B. Demple. 2002. Global adjustment of microbial physiology during free radical stress. Adv. Microb. Physiol. 46:319-341.
8. Keyer, K., and J. A. Imlay. 1996. Superoxide accelerates DNA damage by elevating free-iron levels. Proc. Natl. Acad. Sci. U. S. A. 93:13635-13640.
9. Workentine, M. L., J. J. Harrison, P. U. Stenroos, H. Ceri, and R. J. Turner. 2008. Pseudomonas fluorescens' view of the periodic table. Environ. Microbiol. 10:238-250. doi: 10.1111/j.1462-2920.2007.01448.x.
10. Stadtman, E. R., and R. L. Levine. 2003. Free radical-mediated oxidation of free amino acids and amino acid residues in proteins. Amino Acids. 25:207-218. doi: 10.1007/s00726-003-0011-2.
11. Stadtman, E. R. 1993. Oxidation of free amino acids and amino acid residues in proteins by radiolysis and by metal-catalyzed reactions. Annu. Rev. Biochem. 62:797-821. doi: 10.1146/annurev.bi.62.070193.004053.
12. Macomber, L., and J. A. Imlay. 2009. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc. Natl. Acad. Sci. U. S. A. 106:8344-8349. doi: 10.1073/pnas.0812808106; 10.1073/pnas.0812808106.
13. Xu, H., F. Qu, H. Xu, W. Lai, Y. Andrew Wang, Z. P. Aguilar, and H. Wei. 2012. Role of reactive oxygen species in the antibacterial mechanism of silver nanoparticles on Escherichia coli O157:H7. Biometals. 25:45-53. doi: 10.1007/s10534-011-9482-x; 10.1007/s10534-011-9482-x.
14. Nies, D. H., and S. Silver. 1995. Ion efflux systems involved in bacterial metal resistances. J. Ind. Microbiol. 14:186-199.
15. Holland, S. L., E. Ghosh, and S. V. Avery. 2010. Chromate-induced sulfur starvation and mRNA mistranslation in yeast are linked in a common mechanism of Cr toxicity. Toxicol. in. Vitro. 24:1764-1767. doi: 10.1016/j.tiv.2010.07.006.
16. Hong, R., T. Y. Kang, C. A. Michels, and N. Gadura. 2012. Membrane lipid peroxidation in copper alloy-mediated contact killing of Escherichia coli. Appl. Environ. Microbiol. 78:1776-1784. doi: 10.1128/AEM.07068-11; 10.1128/AEM.07068-11.
17. Dibrov, P., J. Dzioba, K. K. Gosink, and C. C. Hase. 2002. Chemiosmotic Mechanism of Antimicrobial Activity of Ag+ in Vibrio cholerae. Antimicrob. Agents Chemother. 46:2668-2670. doi: 10.1128/AAC.46.8.2668-2670.2002.
18. Warnes, S. L., C. J. Highmore, and C. W. Keevil. 2012. Horizontal Transfer of Antibiotic Resistance Genes on Abiotic Touch Surfaces: Implications for Public Health. mBio. 3:. doi: 10.1128/mBio.00489-12.
19. Xu, K. D., P. S. Stewart, F. Xia, C. T. Huang, and G. A. McFeters. 1998. Spatial Physiological Heterogeneity in Pseudomonas aeruginosa Biofilm Is Determined by Oxygen Availability. Appl. Environ. Microbiol. 64:4035-4039.
20. Harrison, J., H. Ceri, J. Yerly, C. Stremick, Y. Hu, R. Martinuzzi, and R. Turner. 2006. The use of microscopy and three-dimensional visualization to evaluate the structure of microbial biofilms cultivated in the calgary biofilm device. Biol. Proc. Online. 8:194-215. doi: 10.1251/bpo127.
21. Hassett, D. J., J. F. Ma, J. G. Elkins, T. R. McDermott, U. A. Ochsner, S. E. West, C. T. Huang, J. Fredericks, S. Burnett, P. S. Stewart, G. McFeters, L. Passador, and B. H. Iglewski. 1999. Quorum sensing in Pseudomonas aeruginosa controls expression of catalase and superoxide dismutase genes and mediates biofilm susceptibility to hydrogen peroxide. Mol. Microbiol. 34:1082-1093.
22. Cornelis, P. 2008. Unexpected Interaction of a Siderophore with Aluminum and Its Receptor. J. Bacteriol. 190:6541-6543.