Leptospira Interrogans: Difference between revisions
Egratke4272 (talk | contribs) |
Egratke4272 (talk | contribs) |
||
| (39 intermediate revisions by the same user not shown) | |||
| Line 3: | Line 3: | ||
[[File:Leptospira interrogans strain RGA 01.png|450px|thumb|right|Leptospira interrogans strain RGA 01. Photo credit: CDC/Dr. Janice Carr Source:CDC's [http://phil.cdc.gov/phil/home.asp Public Health Image Library] Image #1220 |alt=Leptospira interrogans strain RGA 01.png]] | [[File:Leptospira interrogans strain RGA 01.png|450px|thumb|right|Leptospira interrogans strain RGA 01. Photo credit: CDC/Dr. Janice Carr Source:CDC's [http://phil.cdc.gov/phil/home.asp Public Health Image Library] Image #1220 |alt=Leptospira interrogans strain RGA 01.png]] | ||
<br><i>Leptospira interrogans</i> is a thin, helical shaped bacteria that causes the zoonotic disease known as Leptospirosis. Leptospirosis is considered the most widespread zoonotic disease | <br><i>Leptospira interrogans</i> is a thin, helical shaped bacteria that causes the [http://www.medicinenet.com/script/main/art.asp?articlekey=12958 zoonotic] disease known as [http://en.wikipedia.org/wiki/Leptospirosis Leptospirosis] [2]. Leptospirosis is considered the most widespread zoonotic disease, can be deadly in severe, untreated cases, and is now the world's leading cause of [http://www.dgif.virginia.gov/wildlife/diseases/hd.asp hemorrhagic disease] [1, 13]. The spread of the disease can be attributed to the ability of <i>L. interrogans</i> to thrive in soil and water without a host for extended periods of time [1]. The disease is difficult to prevent in humans due to the high amount of variety in [http://medical-dictionary.thefreedictionary.com/serovar serovars] of <i>L. interrogans</i>, but current research is moving toward the creation of a human vaccination against Leptospirosis and a more indepth understanding of the genome and infectious mechanisms of the species [1]. The wide variety of serovars and host species are challenges that must be overcome, but specific proteins and gene sequences common between serovars are being discovered based on genome analysis, and it is becoming more and more likely that a vaccine to prevent Leptospirosis will be developed [13]. | ||
==Classification== | ==Classification== | ||
| Line 15: | Line 15: | ||
[[File:Leptospira scanning micrograph.jpg|300px|thumb|left|Leptospira under a scanning micrograph. Photo credit: CDC/Rob Weyant Source:CDC's [http://phil.cdc.gov/phil/home.asp Public Health Image Library] Image #138|alt=Leptospira scanning micrograph.jpg]] | [[File:Leptospira scanning micrograph.jpg|300px|thumb|left|Leptospira under a scanning micrograph. Photo credit: CDC/Rob Weyant Source:CDC's [http://phil.cdc.gov/phil/home.asp Public Health Image Library] Image #138|alt=Leptospira scanning micrograph.jpg]] | ||
<br><b>Cellular Biology</b> | |||
<br><i>Leptospira interrogans</i> is a type of gram negative bacteria [2]. The cell is thin and spiral shaped with a hook on each end [2]. It is motile and possesses two periplasmic flagella [2]. | |||
<br><b>Serovars</b> | |||
<br>The <i>L. interrogans</i> species can be broken down into roughly 290 serovars [2]. Leptospiral serovar diversity results from structural differences in carbohydrate component of lipopolysaccharides [4]. Many serovars are adapted for specific mammalian reservoir hosts [4]. Many serovars have structural differences, but their genomes and mechanisms of infection remain extremely similar [4]. | |||
<br>< | <br><b>Metabolism</b> | ||
<i>Leptospira interrogans</i> is an aerobic bacteria and possesses both catalase and oxidase enzymes and does not ferment carbohydrates [2]. <i>Leptospira</i> species have only one glucose uptake system [4]. This system is a [http://en.wikipedia.org/wiki/Sodium-glucose_transport_proteins glucose-sodium symporter] that is dependent on a sodium gradient across the bacterial membrane [4]. This limited system results in an inability to utilize glucose as an energy source in many environments [4]. Consequently, <i>Leptospira</i> species utilize beta-oxidation of long-chain fatty acids as the major energy and carbon source instead of the more common sugar oxidative pathways [4]. | |||
==Genomic Structure== | ==Genomic Structure== | ||
<br>The <i>L. interrogans</i> genome consists of 4,691,184 base pairs (bp). The genome consists of two circular chromosomes: a larger one of 4,332,241 bp (CI) and a smaller one of 358,943 bp [8]. Unlike most other bacteria, rRNA genes in <i>L. interrogans</i> are not organized in operons; they are scattered over the CI chromosome. Signal transduction mechanisms are regulated by at least 79 genes encoding two-component sensor histidine kinase-response regulator proteins. This allows <i>L. interrogans</i> to respond to a wide variety of environmental factors. Two copies of genes encoding BolA-like proteins can be identified in the <i>L. interrogans</i> | <br><b>Overview</b> | ||
<br>The <i>L. interrogans</i> genome consists of 4,691,184 base pairs (bp) [8]. The genome consists of two circular chromosomes: a larger one of 4,332,241 bp (CI) and a smaller one of 358,943 bp [8]. Unlike most other bacteria, rRNA genes in <i>L. interrogans</i> are not organized in operons; they are scattered over the CI chromosome [7]. Signal transduction mechanisms are regulated by at least 79 genes encoding two-component sensor histidine kinase-response regulator proteins [7]. This allows <i>L. interrogans</i> to respond to a wide variety of environmental factors [7]. The <i>L. interrogans</i> genome contains at least 263 genes that encode potential surface-exposed integral membrane proteins [4]. At least 79 of the 4768 predicted genes identified in the genome sequence are related to motility [4]. Two copies of genes encoding BolA-like proteins can be identified in the <i>L. interrogans</i> genome but not in the genomes of two other spirochetes, [http://en.wikipedia.org/wiki/Treponema_pallidum <i>Treponema pallidum</i>] and [http://en.wikipedia.org/wiki/Borrelia_burgdorferi <i>Borrelia burgdorferi</i>]. BolA has recently been linked to maintenance of cell shape in extreme conditions, suggesting that <i>L. interrogans</i> has this capability [7]. Only 37 of the genes encoded within the genome code for tRNA [8]. This number is extremely low and is believed to be one of the factors attributing to the fastidious growth of the pathogen [8]. | |||
[[File:Microbe Wiki Figure.JPG|600px|thumb|left|Figure 1. Genomic feature comparison of <i>L. interrogans</i> serovars Copenhageni and Lai. Data is provided courtesy of Nascimento et al. http://jb.asm.org/content/186/7/2164.full (See reference 4 below)]] | |||
<br>< | <br><b>Serovar Differences</b> | ||
<br>The Lai serovar is the most heavily researched and studied, but in recent years, various serovars have been sequenced and compared to the readily available Lai genomic information [4]. Despite structural differences such as chromosomal inversion and rearrangement of many insertion elements, serovars Copenhageni and Lai are genetically similar [4]. Specific similarities and differences between sequence numbers and lengths can be viewed in the figure to the left (Figure 1). Both Copenhageni and Lai have 79 genes dedicated to motility, and both possess five genes encoding secreted enzymes used to break down the preliminary defenses of host cells [4]. Additionally, there are two families of [http://en.wikipedia.org/wiki/Bacterial_adhesin afimbrial adhesins] not previously described for Lai that can be described for Copenhageni, and these may contribute to the early steps of infection and colonization of host tissue [4]. Differences between the two serovars can likely be attributed to differences in their primary hosts: [http://en.wikipedia.org/wiki/Brown_rat <i>Rattus norvegicus</i>] in serovar Copenhageni and [http://en.wikipedia.org/wiki/Striped_field_mouse <i>Apodemus agrarius</i>] in serovar Lai [4]. The information provided by genomic comparisons of various <i>L. interrogans</i> serovars indicates that while serovars can differ morphologically due to different host environments, many of the genetic make ups and their subsequent virulence mechanisms are similar across serovars [4]. | |||
<br><b>Genes of Particular Interest</b> | |||
<br><b> | <br>The genome of L. interrogans encodes several proteins bearing homology to animal proteins important in [http://en.wikipedia.org/wiki/Hemostasis hemostasis] [8]. One of these is resembles mammalian platelet-activating factor (PAF) [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2674695/ acetylhydrolase14] and another is similar to [http://en.wikipedia.org/wiki/Von_Willebrand_factor von Willebrand factor15] type A domains [8]. No bacterial genomes have yet been shown to encode both of these proteins, but several bacterial species possess them individually [8]. A third gene relevant to hemostasis, so far found only in Leptospira, seems to specify an orthologue of [http://en.wikipedia.org/wiki/Paraoxonase paraoxonase]. These genes, among others, code for proteins believed to help break down external matrices and membranes of host cells to better facilitate invasian [7]. <i>L. interrogans</i> possesses several genes related to the attachment and invasion of eukaryotic cells: mce, invA, atsE and mviN [8]. The 79 motility genes mentioned in the above overview also contribute to the pathogenesis of <i>L. interrogans</i> [8]. These genes, along with the complete set that determine the shape of the bacteria, give rise to physical attributes (hooks, multiple flagella, etc.) that are pathogenically advantageous and lead to more effective and efficient colonization of host cells [4]. Another virulence factor of <i>L. interrogans</i> , [http://en.wikipedia.org/wiki/Chemotaxis chemotaxis], is coded for by a high number of genes (12 coding sequences) [8]. This again indicates that <i>L. interrogans</i> can exist in a wide variety of environmental conditions, which partially explains its prominence in so many mammalian reservoir species [8]. Additionally, 12 L. interrogans genes that encode proteins with 27- to 33-amino-acid ankyrin repeat domains have been identified [4]. In prokaryotes, these genes have been found located near genes involved in either nutrient acquisition or tolerance/resistance to antibiotics[4]. In all, there are an estimated 366 genes in <i>L. interrogans</i> that are related to virulence and pathogenic factors [4]. | ||
==Pathogenesis== | |||
<br> | <br><b>Infection</b> | ||
<br><i>L. interrogans</i> can be spread through the bodily fluids, excluding the saliva, of infected animals [1]. The bacteria can enter the body through skin or mucous membranes and via consumption of contaminated water [1]. Infected wild and domestic animals may continue to excrete the bacteria into the environment for up to several years and the bacteria can remain in soil and water for months at a time [1]. The ability of the pathogen to remain viable in soil and water supplies makes it extremely dangerous and prevalent in developing nations where food and water sources are not always sanitized prior to consumption [7]. Once the bacteria has been ingested, virulence mechanisms such as motility and chemotaxis responses that are extensively coded for the in the genome enable bacteria to penetrate the host tissues rapidly [8]. Motility contributes greatly to the decimating effect of the disease from the site of entry to sites of end-organ damage in the lungs, liver, kidney, eyes, and brain [7]. The flagella and shape of the cells allow them to move through the body and the hooks on the ends of their cells allow them to attach and latch on to host tissues [7]. After reaching the blood stream, targeted sites of colonization are the liver and kidneys [11]. These organs have a large supply of lipids that are essential for the growth and survival of <i>L. interrogans</i> [11]. <i>L. interrogans</i> tends to bind to a variety of cell lines including fibroblasts, endothelial cells, and kidney epithelial cells [11]. Once the bacteria attach to the host cell, <i>L. interrogans</i> cells secrete enzymes that degrade host cell matrices, allowing for faster invasion and infection [4]. The bacteria reproduce within the cell and are released slowly into the surrounding environmnt while waiting for the immune system to mount a response [12]. Virulent leptospiras can protect themselves against phagocytic cells and the complement system [11]. Pathogenic leptospiras escape from phagocytosis, are resistant to intracellular killing mechanisms and evade the complement system by binding to complement system inhibitor FH [11]. FH is a regulatory complement protein that prevents complement activation and binding to it can restrict the deposition of the late complement components on bacteria surfaces [11]. Once the host immune system does kill the bacteria, the bacteria release [http://textbookofbacteriology.net/endotoxin.html endotoxins] [11]. The innate immune system recognizes endotoxins through specific receptors and mediates a response by [http://en.wikipedia.org/wiki/Toll-like_receptor Toll-like receptors] and [http://en.wikipedia.org/wiki/Na%2B/K%2B-ATPase Na+/K+-ATPase] [11]. These sense antigen molecules and trigger intracellular signaling pathways driving the translocation of transcription factors which lead to increased inflammatory mediator production, creating an inflammatory microenvironment that can lead to organ dysfunction [11]. After infection, acquired immunity does sometimes occur, but it seems to be dependent on the specific serovar of infection [11]. Patients who have recovered from leptospirosis do not seem to generate memory T cells that can be activated by in vitro stimulation with Leptospiral protein antigens [11]. | |||
[[File:Leptospirosis in kidney.jpg|300px|thumb|right|Photomicrograph of Leptospira bacteria in kidney tissue. Photo credit: CDC/Dr. Martin Hicklin Source:CDC's [http://phil.cdc.gov/phil/home.asp Public Health Image Library] Image #2769|alt=Leptospirosis in kidney.jpg]] | |||
<br> | <br><b>Presentation in Humans</b> | ||
<br>Human infection by <i>L. interrogans</i> is classified as either [http://www.merriam-webster.com/dictionary/icteric icteric] or [http://www.merriam-webster.com/medical/anicteric anicteric] [1]. The anticteric form makes up 90% of infections and is less severe [1]. The icteric form makes up 10% of infections and is very severe. The icteric form is also known as [http://www.healthline.com/health/weils-disease#Symptoms4 Weil’s Disease] [1]. Weil’s Disease is most often due to infection with serovar [https://www.zoetisus.com/conditions/beef/leptospira-icterohaemorrhagiae.aspx <i>L. icterohaemorrhagiae</i>], but humans are susceptible to infection via a variety of serovars [3]. Typically, symptoms begin to appear within two to four weeks of exposure [1]. The infection can be divided into two phases: phase one which represents the anticteric infection, and phase two which represents the icteric infection [1]. In phase one, symptoms include fever, chills, headache, muscle aches, vomiting, or diarrhea [1]. These symptoms can often be mistaken for other infections, and in some cases, no symptoms appear [1]. If the infection progresses into phase two (Weil’s disease), symptoms include [http://en.wikipedia.org/wiki/Petechia petechiae], [http://www.nlm.nih.gov/medlineplus/ency/article/003275.htm hepatomegaly], [http://en.wikipedia.org/wiki/Jaundice jaundice], renal tubular damage and subsequent renal insufficiency [4]. The disease can progress and affect almost all internal organs, producing inflammation and hepatic lesions that lead to organ failure [4]. If the infection and liver/renal failure are not promptly diagnosed and treated, recovery can take months, and mortality can reach 20% [3]. | |||
<br><b>Other Mammalian Presentation</b> | |||
<br> | <br>During the first 4-12 days following infection, symptoms include fever, depression, vomiting, loss of appetite, [http://www.webmd.com/eye-health/eye-health-conjunctivitis conjunctivitis], and generalized pain [9]. Within 2 days of the onset of these primary symptoms, body temperature may drop suddenly and there may be a noticeable increase in thirst [9]. Color intensity of the urine may vary from lemon to deep orange [10]. This color change combined with jaundice can be the only diagnostic indicators of Leptospirosis [10]. Additionally, frequent urination and subsequent dehydration (uremia) are consistent with invasion of the kidney tubule cells by <i>L. interrogans</i> [10]. In advanced cases of infection, difficulty breathing, muscular tremors, and bloody excrement are often observed [9]. These symptoms appear as the infection progresses to include the liver, gastrointestinal system and other organs [9]. Course and severity of the disease is often dependent upon the serovar responsible for the infection [9]. | ||
== | ==Prevention and Ongoing Research== | ||
<br><b>Prevention</b> | |||
<br>Due to the common transmission of <i>L. interrogans</i> via rodents, one of the methods of prevention in animals is minimizing rodent populations near animal and human residences [1]. This can be accomplished by using pest control methods, but it is near impossible in many areas to restrict rodent and animal access to shared human resources. Pets can be vaccinated against Leptospirosis, but the vaccine only inhibits a handful of serovars and vaccination must be repeated annually [1]. There is no formal method of Leptospirosis prevention in humans [1]. Chance of contraction can be reduced by minimizing or avoiding direct contact with animal excrement or water contaminated with animal waste [1]. | |||
<br>The vaccines currently available to target <i>L. interrogans</i> have low efficacy, are serovar-specific, and do not induce long-term protection against infection. Major limitations to improvements include the large number of pathogenic serovars and the cost of producing a multiserovar vaccine. It is anticpated that examination of candidate protective immunogens, such as conserved outer membrane proteins among serovars will provide new approaches for vaccine development [4]. In | <br><b>Vaccine Development</b> | ||
<br>The vaccines currently available to target <i>L. interrogans</i> have low [http://en.wikipedia.org/wiki/Efficacy efficacy], are serovar-specific, and do not induce long-term protection against infection [4]. Major limitations to improvements include the large number of pathogenic serovars and the cost of producing a multiserovar vaccine [4]. It is anticpated that examination of candidate protective immunogens, such as conserved outer membrane proteins among serovars will provide new approaches for vaccine development [4]. In a study conducted 2005, this approach was taken, and sixteen proteins, out of a hundred tested, were recognized by antibodies present in human sera [5]. Four of these proteins were conserved among eight serovars of <i>L. interrogans</i> [5]. These proteins could be used to develop a more effective vaccination [5]. Surface proteins are likely to be the key to developing a multiserovar vaccine for Leptospirosis [5]. Over the course of the last few years, a new technique to identify vaccine candidates has been utilized: [http://en.wikipedia.org/wiki/Reverse_vaccinology reverse vaccinology] [13]. This approach, using [http://en.wikipedia.org/wiki/Bioinformatics bioinformatics], narrows down the number of potential vaccine antigen candidates by identifying [http://en.wikipedia.org/wiki/Proteomics proteomic] features possibly associated with [http://en.wikipedia.org/wiki/Antigenicity antigenicity] and vaccine efficacy based on the amino acid sequence alone, reducing the number of candidates to several hundred [13]. The information provided by reverse vaccinology is useful, but it still necessitates a a large amount of research to explore the resulting several hundred candidates [13]. Bioinformatics can also often be simplistic and miss crucial antigens [13]. The method has still been used successfully, and in a study conducted in 2014, 12 proteins conserved between 5 leptospiral genomes were identified as potential vaccine candidates [14]. Additionally, the study that obtained these results used whole genome analysis rather than selected parts, and also included non-classical proteins in their search, resulting in what could be a more complete set of viable vaccine proteins [14]. Biological confirmation of the 12 derived proteins would still need to be conducted to confirm the results, but the information from the study and others like it is very promising [14]. A different method of research that is being explored is the use of [http://en.wikipedia.org/wiki/Protein_microarray proteome microarray chips] [13]. Proteomic studies are useful in that they use human serum to identify proteins that are used primarily in mechanisms of infection [13]. A study conducted in 2014 utilized this method, and tested 188 human serum samples from patients with varying degrees of Leptospirosis presentation [13]. Forty-nine of the discovered 191 reactive antigens were reactive for both IgM and IgG antibodies, and the recognition of these antigens provides a large amount of insight into the mechanism of immune response against leptospiosis [13]. Understanding of this immune response could lead to a better approach to developing an effective vaccine [13]. Further sequencing and analysis of serovar genomes has aided in the development of a greater understanding of Leptospirosis caused by <i>L. interrogans</i>, but there is still much to be discovered regarding this pathogen, and researchers around the world are working diligently to add to the literature surrounding it so that we may more readily and effectively prevent and treat Leptospirosis. | |||
==Further Reading== | ==Further Reading== | ||
| Line 53: | Line 66: | ||
<br>4. [http://jb.asm.org/content/186/7/2164.full Nascimento, A. L. T. O. et al. “Comparative Genomics of Two Leptospira Interrogans Serovars Reveals Novel Insights into Physiology and Pathogenesis .” Journal of Bacteriology 186.7 (2004): 2164–2172. PMC. Web. 26 Feb. 2015.] | <br>4. [http://jb.asm.org/content/186/7/2164.full Nascimento, A. L. T. O. et al. “Comparative Genomics of Two Leptospira Interrogans Serovars Reveals Novel Insights into Physiology and Pathogenesis .” Journal of Bacteriology 186.7 (2004): 2164–2172. PMC. Web. 26 Feb. 2015.] | ||
<br>5. [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1949541/pdf/amjpathol00329-0024.pdf Arean, Victor M., M.D. "The Pathologic Anatomy and Pathogenesis of Fatal Human Leptospirosis (Weil's Disease)." The American Journal of Pathology 40.4 (1962): 393- 423. Web. 25 Feb. 2015.] | <br>5. [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1949541/pdf/amjpathol00329-0024.pdf Arean, Victor M., M.D. "The Pathologic Anatomy and Pathogenesis of Fatal Human Leptospirosis (Weil's Disease)." The American Journal of Pathology 40.4 (1962): 393- 423. Web. 25 Feb. 2015.] | ||
<br>6. [http://femsle.oxfordjournals.org/content/244/2/305. | <br>6. [http://femsle.oxfordjournals.org/content/244/2/305.long Gamberini, Marcia, Ricardo M. Gomez, Marina V. Atzingen, Elizabeth AL Martins, Silvio A. Vasconcellos, Eliete C. Romero, Luciana C.C. Leite, Paulo L. Ho, and Ana L.T.O Nascimento. "Whole-genome Analysis of Leptospira Interrogans to Identify Potential Vaccine Candidates against Leptospirosis." FEMS Microbiology Letters 244.2 (2005): 305-13. Web. 24 Feb. 2015.] | ||
<br>7. [http://www. | <br>7. [http://www.sciencedirect.com/science/article/pii/S1473309903008302 Bharti, Ajay R., Jarlath E. Nally, Jessica N. Ricaldi, Michael A. Matthias, Monica M. Diaz, Michael A. Lovett, Paul N. Levett, Robert H. Gilman, Michael R. Willig, Eduardo Gotuzzo, and Joseph M. Vinetz. "Leptospirosis: A Zoonotic Disease of Global Importance." The Lancet Infectious Diseases 3.12 (2003): 757-71. Web. 25 Feb. 2015.] | ||
<br>8. [http://www.nature.com/nature/journal/v422/n6934/full/nature01597.html Ren, Shuang-Xi, Gang Fu, Xiu-Gao Jiang, Rong Zeng, You-Gang Miao, Hai Xu, Yi-Xuan Zhang, Hui Xiong, Gang Lu, Ling-Feng Lu, Hong-Quan Jiang, Jia Jia, Yue-Feng Tu, Ju-Xing Jiang, Wen-Yi Gu, Yue-Qing Zhang, Zhen Cai, Hai-Hui Sheng, Hai-Feng Yin, Yi Zhang, Gen-Feng Zhu, Ma Wan, Hong-Lei Huang, Zhen Qian, Sheng-Yue Wang, Wei Ma, Zhi-Jian Yao, Yan Shen, Bo-Qin Qiang, Qi-Chang Xia, Xiao-Kui Guo, Antoine Danchin, Isabelle Saint Girons, Ronald L. Somerville, Yu-Mei Wen, Man-Hua Shi, Zhu Chen, Jian-Guo Xu, and Guo-Ping Zhao. "Unique Physiological and Pathogenic Features of Leptospira Interrogans Revealed by Whole-genome Sequencing." Nature 422.6934 (2003): 888-93. Nature. Web. 23 Mar. 2015.] | <br>8. [http://www.nature.com/nature/journal/v422/n6934/full/nature01597.html Ren, Shuang-Xi, Gang Fu, Xiu-Gao Jiang, Rong Zeng, You-Gang Miao, Hai Xu, Yi-Xuan Zhang, Hui Xiong, Gang Lu, Ling-Feng Lu, Hong-Quan Jiang, Jia Jia, Yue-Feng Tu, Ju-Xing Jiang, Wen-Yi Gu, Yue-Qing Zhang, Zhen Cai, Hai-Hui Sheng, Hai-Feng Yin, Yi Zhang, Gen-Feng Zhu, Ma Wan, Hong-Lei Huang, Zhen Qian, Sheng-Yue Wang, Wei Ma, Zhi-Jian Yao, Yan Shen, Bo-Qin Qiang, Qi-Chang Xia, Xiao-Kui Guo, Antoine Danchin, Isabelle Saint Girons, Ronald L. Somerville, Yu-Mei Wen, Man-Hua Shi, Zhu Chen, Jian-Guo Xu, and Guo-Ping Zhao. "Unique Physiological and Pathogenic Features of Leptospira Interrogans Revealed by Whole-genome Sequencing." Nature 422.6934 (2003): 888-93. Nature. Web. 23 Mar. 2015.] | ||
<br>9. [http://www.labbies.com/lepto.htm Davol, Pamela A. "Canine Leptospirosis." Wing-N-Wave Labradors. N.p., n.d. Web. 24 Mar. 2015.] | <br>9. [http://www.labbies.com/lepto.htm Davol, Pamela A. "Canine Leptospirosis." Wing-N-Wave Labradors. N.p., n.d. Web. 24 Mar. 2015.] | ||
<br>10. [http://onlinelibrary.wiley.com/doi/10.1111/j.1748-5827.1998.tb03640.x/abstract Birnbaum, N., S. C. Barr, S. A. Center, T. Schermerhorn, J. F. Randolph, and K. W. Simpson. "Naturally Acquired Leptospirosis in 36 Dogs: Serological and Clinicopathological Features." Journal of Small Animal Practice 39.5 (1998): 231-36. Web. 24 Feb. 2015.] | <br>10. [http://onlinelibrary.wiley.com/doi/10.1111/j.1748-5827.1998.tb03640.x/abstract Birnbaum, N., S. C. Barr, S. A. Center, T. Schermerhorn, J. F. Randolph, and K. W. Simpson. "Naturally Acquired Leptospirosis in 36 Dogs: Serological and Clinicopathological Features." Journal of Small Animal Practice 39.5 (1998): 231-36. Web. 24 Feb. 2015.] | ||
<br>11. [http://www.hindawi.com/journals/mi/2012/317950/ Gonçalves-De-Albuquerque, C. F., P. Burth, A. R. Silva, M. Younes-Ibrahim, H. C. Castro-Faria-Neto, and M. V. Castro-Faria. "Leptospira and Inflammation." Mediators of Inflammation: 1-11. Web. 11 April 2015.] | |||
<br>12. [http://onlinelibrary.wiley.com/doi/10.1111/j.1462-5822.2011.01660.x/full Toma, Claudia, Nobuhiko Okura, Chitoshi Takayama, and Toshihiko Suzuki. "Characteristic Features of Intracellular Pathogenic Leptospira in Infected Murine Macrophages." Cellular Microbiology: 1783-792. Web. 11 April 2015.] | |||
<br>13. [http://ry6af4uu9w.search.serialssolutions.com/?&url_ver=Z39.88-2004&url_ctx_fmt=info:ofi/fmt:kev:mtx:ctx&rft_val_fmt=info:ofi/fmt:kev:mtx:journal&rft.atitle=Proteomic%20Features%20Predict%20Seroreactivity%20against%20Leptospiral%20Antigens%20in%20Leptospirosis%20Patients&rft.aufirst=Carolina&rft.aulast=Lessa-Aquino&rft.date=2015&rft.eissn=1535-3907&rft.epage=556&rft.genre=article&rft.issn=1535-3893&rft.issue=1&rft.jtitle=JOURNAL%20OF%20PROTEOME%20RESEARCH&rft.pages=549-556&rft.spage=549&rft.stitle=J%20PROTEOME%20RES&rft.volume=14&rfr_id=info:sid/www.isinet.com:WoK:WOS&rft.au=Wunder%2C%20Elsio%20A%2E%2C%20Jr%2E&rft.au=Lindow%2C%20Janet%20C%2E&rft.au=Rodrigues%2C%20Camila%20B%2E&rft.au=Pablo%2C%20Jozelyn&rft_id=info:pmid/25358092&rft_id=info:doi/10%2E1021%2Fpr500718t Lessa-Aquino, Carolina, Elsio A. Wunder, Janet C. Lindow, Camila B. Rodrigues, Jozelyn Pablo, Rie Nakajima, Algis Jasinskas, Li Liang, Mitermayer G. Reis, Albert I. Ko, Marco A. Medeiros, and Philip L. Felgner. "Proteomic Features Predict Seroreactivity against Leptospiral Antigens in Leptospirosis Patients." Journal of Proteome Research (2014). American Chemical Society. Web. 10 Apr. 2015.] | |||
<br>14. [http://link.springer.com/article/10.1007/s12010-015-1507-4/fulltext.html Mudadu, Mauricio De Alvarenga, Viviane Carvalho, and Sophie Y. Leclercq. "Nonclassically Secreted Proteins as Possible Antigens for Vaccine Development: A Reverse Vaccinology Approach." Applied Biochemistry and Biotechnology (2015): n. pag. 12 Feb. 2015. Web. 11 Apr. 2015.] | |||
<!--Do not remove this line--> | <!--Do not remove this line--> | ||
Latest revision as of 17:29, 14 April 2015
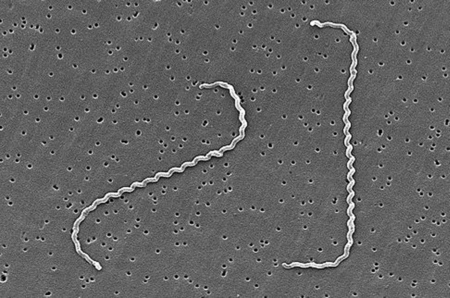
Leptospira interrogans is a thin, helical shaped bacteria that causes the zoonotic disease known as Leptospirosis [2]. Leptospirosis is considered the most widespread zoonotic disease, can be deadly in severe, untreated cases, and is now the world's leading cause of hemorrhagic disease [1, 13]. The spread of the disease can be attributed to the ability of L. interrogans to thrive in soil and water without a host for extended periods of time [1]. The disease is difficult to prevent in humans due to the high amount of variety in serovars of L. interrogans, but current research is moving toward the creation of a human vaccination against Leptospirosis and a more indepth understanding of the genome and infectious mechanisms of the species [1]. The wide variety of serovars and host species are challenges that must be overcome, but specific proteins and gene sequences common between serovars are being discovered based on genome analysis, and it is becoming more and more likely that a vaccine to prevent Leptospirosis will be developed [13].
Classification
Higher Order Taxa
Bacteria; Spirochaetes; Spirochaetia; Spirochaetales; Leptospiraceae; Leptospira
Species
Leptospira interrogans
Cell Structure and Metabolism
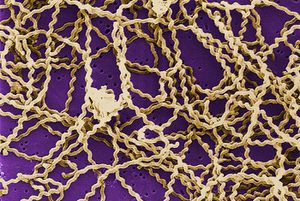
Cellular Biology
Leptospira interrogans is a type of gram negative bacteria [2]. The cell is thin and spiral shaped with a hook on each end [2]. It is motile and possesses two periplasmic flagella [2].
Serovars
The L. interrogans species can be broken down into roughly 290 serovars [2]. Leptospiral serovar diversity results from structural differences in carbohydrate component of lipopolysaccharides [4]. Many serovars are adapted for specific mammalian reservoir hosts [4]. Many serovars have structural differences, but their genomes and mechanisms of infection remain extremely similar [4].
Metabolism
Leptospira interrogans is an aerobic bacteria and possesses both catalase and oxidase enzymes and does not ferment carbohydrates [2]. Leptospira species have only one glucose uptake system [4]. This system is a glucose-sodium symporter that is dependent on a sodium gradient across the bacterial membrane [4]. This limited system results in an inability to utilize glucose as an energy source in many environments [4]. Consequently, Leptospira species utilize beta-oxidation of long-chain fatty acids as the major energy and carbon source instead of the more common sugar oxidative pathways [4].
Genomic Structure
Overview
The L. interrogans genome consists of 4,691,184 base pairs (bp) [8]. The genome consists of two circular chromosomes: a larger one of 4,332,241 bp (CI) and a smaller one of 358,943 bp [8]. Unlike most other bacteria, rRNA genes in L. interrogans are not organized in operons; they are scattered over the CI chromosome [7]. Signal transduction mechanisms are regulated by at least 79 genes encoding two-component sensor histidine kinase-response regulator proteins [7]. This allows L. interrogans to respond to a wide variety of environmental factors [7]. The L. interrogans genome contains at least 263 genes that encode potential surface-exposed integral membrane proteins [4]. At least 79 of the 4768 predicted genes identified in the genome sequence are related to motility [4]. Two copies of genes encoding BolA-like proteins can be identified in the L. interrogans genome but not in the genomes of two other spirochetes, Treponema pallidum and Borrelia burgdorferi. BolA has recently been linked to maintenance of cell shape in extreme conditions, suggesting that L. interrogans has this capability [7]. Only 37 of the genes encoded within the genome code for tRNA [8]. This number is extremely low and is believed to be one of the factors attributing to the fastidious growth of the pathogen [8].
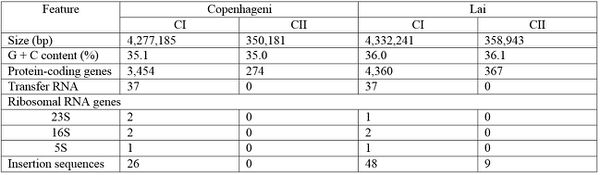
Serovar Differences
The Lai serovar is the most heavily researched and studied, but in recent years, various serovars have been sequenced and compared to the readily available Lai genomic information [4]. Despite structural differences such as chromosomal inversion and rearrangement of many insertion elements, serovars Copenhageni and Lai are genetically similar [4]. Specific similarities and differences between sequence numbers and lengths can be viewed in the figure to the left (Figure 1). Both Copenhageni and Lai have 79 genes dedicated to motility, and both possess five genes encoding secreted enzymes used to break down the preliminary defenses of host cells [4]. Additionally, there are two families of afimbrial adhesins not previously described for Lai that can be described for Copenhageni, and these may contribute to the early steps of infection and colonization of host tissue [4]. Differences between the two serovars can likely be attributed to differences in their primary hosts: Rattus norvegicus in serovar Copenhageni and Apodemus agrarius in serovar Lai [4]. The information provided by genomic comparisons of various L. interrogans serovars indicates that while serovars can differ morphologically due to different host environments, many of the genetic make ups and their subsequent virulence mechanisms are similar across serovars [4].
Genes of Particular Interest
The genome of L. interrogans encodes several proteins bearing homology to animal proteins important in hemostasis [8]. One of these is resembles mammalian platelet-activating factor (PAF) acetylhydrolase14 and another is similar to von Willebrand factor15 type A domains [8]. No bacterial genomes have yet been shown to encode both of these proteins, but several bacterial species possess them individually [8]. A third gene relevant to hemostasis, so far found only in Leptospira, seems to specify an orthologue of paraoxonase. These genes, among others, code for proteins believed to help break down external matrices and membranes of host cells to better facilitate invasian [7]. L. interrogans possesses several genes related to the attachment and invasion of eukaryotic cells: mce, invA, atsE and mviN [8]. The 79 motility genes mentioned in the above overview also contribute to the pathogenesis of L. interrogans [8]. These genes, along with the complete set that determine the shape of the bacteria, give rise to physical attributes (hooks, multiple flagella, etc.) that are pathogenically advantageous and lead to more effective and efficient colonization of host cells [4]. Another virulence factor of L. interrogans , chemotaxis, is coded for by a high number of genes (12 coding sequences) [8]. This again indicates that L. interrogans can exist in a wide variety of environmental conditions, which partially explains its prominence in so many mammalian reservoir species [8]. Additionally, 12 L. interrogans genes that encode proteins with 27- to 33-amino-acid ankyrin repeat domains have been identified [4]. In prokaryotes, these genes have been found located near genes involved in either nutrient acquisition or tolerance/resistance to antibiotics[4]. In all, there are an estimated 366 genes in L. interrogans that are related to virulence and pathogenic factors [4].
Pathogenesis
Infection
L. interrogans can be spread through the bodily fluids, excluding the saliva, of infected animals [1]. The bacteria can enter the body through skin or mucous membranes and via consumption of contaminated water [1]. Infected wild and domestic animals may continue to excrete the bacteria into the environment for up to several years and the bacteria can remain in soil and water for months at a time [1]. The ability of the pathogen to remain viable in soil and water supplies makes it extremely dangerous and prevalent in developing nations where food and water sources are not always sanitized prior to consumption [7]. Once the bacteria has been ingested, virulence mechanisms such as motility and chemotaxis responses that are extensively coded for the in the genome enable bacteria to penetrate the host tissues rapidly [8]. Motility contributes greatly to the decimating effect of the disease from the site of entry to sites of end-organ damage in the lungs, liver, kidney, eyes, and brain [7]. The flagella and shape of the cells allow them to move through the body and the hooks on the ends of their cells allow them to attach and latch on to host tissues [7]. After reaching the blood stream, targeted sites of colonization are the liver and kidneys [11]. These organs have a large supply of lipids that are essential for the growth and survival of L. interrogans [11]. L. interrogans tends to bind to a variety of cell lines including fibroblasts, endothelial cells, and kidney epithelial cells [11]. Once the bacteria attach to the host cell, L. interrogans cells secrete enzymes that degrade host cell matrices, allowing for faster invasion and infection [4]. The bacteria reproduce within the cell and are released slowly into the surrounding environmnt while waiting for the immune system to mount a response [12]. Virulent leptospiras can protect themselves against phagocytic cells and the complement system [11]. Pathogenic leptospiras escape from phagocytosis, are resistant to intracellular killing mechanisms and evade the complement system by binding to complement system inhibitor FH [11]. FH is a regulatory complement protein that prevents complement activation and binding to it can restrict the deposition of the late complement components on bacteria surfaces [11]. Once the host immune system does kill the bacteria, the bacteria release endotoxins [11]. The innate immune system recognizes endotoxins through specific receptors and mediates a response by Toll-like receptors and Na+/K+-ATPase [11]. These sense antigen molecules and trigger intracellular signaling pathways driving the translocation of transcription factors which lead to increased inflammatory mediator production, creating an inflammatory microenvironment that can lead to organ dysfunction [11]. After infection, acquired immunity does sometimes occur, but it seems to be dependent on the specific serovar of infection [11]. Patients who have recovered from leptospirosis do not seem to generate memory T cells that can be activated by in vitro stimulation with Leptospiral protein antigens [11].
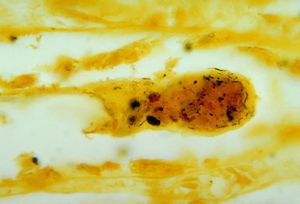
Presentation in Humans
Human infection by L. interrogans is classified as either icteric or anicteric [1]. The anticteric form makes up 90% of infections and is less severe [1]. The icteric form makes up 10% of infections and is very severe. The icteric form is also known as Weil’s Disease [1]. Weil’s Disease is most often due to infection with serovar L. icterohaemorrhagiae, but humans are susceptible to infection via a variety of serovars [3]. Typically, symptoms begin to appear within two to four weeks of exposure [1]. The infection can be divided into two phases: phase one which represents the anticteric infection, and phase two which represents the icteric infection [1]. In phase one, symptoms include fever, chills, headache, muscle aches, vomiting, or diarrhea [1]. These symptoms can often be mistaken for other infections, and in some cases, no symptoms appear [1]. If the infection progresses into phase two (Weil’s disease), symptoms include petechiae, hepatomegaly, jaundice, renal tubular damage and subsequent renal insufficiency [4]. The disease can progress and affect almost all internal organs, producing inflammation and hepatic lesions that lead to organ failure [4]. If the infection and liver/renal failure are not promptly diagnosed and treated, recovery can take months, and mortality can reach 20% [3].
Other Mammalian Presentation
During the first 4-12 days following infection, symptoms include fever, depression, vomiting, loss of appetite, conjunctivitis, and generalized pain [9]. Within 2 days of the onset of these primary symptoms, body temperature may drop suddenly and there may be a noticeable increase in thirst [9]. Color intensity of the urine may vary from lemon to deep orange [10]. This color change combined with jaundice can be the only diagnostic indicators of Leptospirosis [10]. Additionally, frequent urination and subsequent dehydration (uremia) are consistent with invasion of the kidney tubule cells by L. interrogans [10]. In advanced cases of infection, difficulty breathing, muscular tremors, and bloody excrement are often observed [9]. These symptoms appear as the infection progresses to include the liver, gastrointestinal system and other organs [9]. Course and severity of the disease is often dependent upon the serovar responsible for the infection [9].
Prevention and Ongoing Research
Prevention
Due to the common transmission of L. interrogans via rodents, one of the methods of prevention in animals is minimizing rodent populations near animal and human residences [1]. This can be accomplished by using pest control methods, but it is near impossible in many areas to restrict rodent and animal access to shared human resources. Pets can be vaccinated against Leptospirosis, but the vaccine only inhibits a handful of serovars and vaccination must be repeated annually [1]. There is no formal method of Leptospirosis prevention in humans [1]. Chance of contraction can be reduced by minimizing or avoiding direct contact with animal excrement or water contaminated with animal waste [1].
Vaccine Development
The vaccines currently available to target L. interrogans have low efficacy, are serovar-specific, and do not induce long-term protection against infection [4]. Major limitations to improvements include the large number of pathogenic serovars and the cost of producing a multiserovar vaccine [4]. It is anticpated that examination of candidate protective immunogens, such as conserved outer membrane proteins among serovars will provide new approaches for vaccine development [4]. In a study conducted 2005, this approach was taken, and sixteen proteins, out of a hundred tested, were recognized by antibodies present in human sera [5]. Four of these proteins were conserved among eight serovars of L. interrogans [5]. These proteins could be used to develop a more effective vaccination [5]. Surface proteins are likely to be the key to developing a multiserovar vaccine for Leptospirosis [5]. Over the course of the last few years, a new technique to identify vaccine candidates has been utilized: reverse vaccinology [13]. This approach, using bioinformatics, narrows down the number of potential vaccine antigen candidates by identifying proteomic features possibly associated with antigenicity and vaccine efficacy based on the amino acid sequence alone, reducing the number of candidates to several hundred [13]. The information provided by reverse vaccinology is useful, but it still necessitates a a large amount of research to explore the resulting several hundred candidates [13]. Bioinformatics can also often be simplistic and miss crucial antigens [13]. The method has still been used successfully, and in a study conducted in 2014, 12 proteins conserved between 5 leptospiral genomes were identified as potential vaccine candidates [14]. Additionally, the study that obtained these results used whole genome analysis rather than selected parts, and also included non-classical proteins in their search, resulting in what could be a more complete set of viable vaccine proteins [14]. Biological confirmation of the 12 derived proteins would still need to be conducted to confirm the results, but the information from the study and others like it is very promising [14]. A different method of research that is being explored is the use of proteome microarray chips [13]. Proteomic studies are useful in that they use human serum to identify proteins that are used primarily in mechanisms of infection [13]. A study conducted in 2014 utilized this method, and tested 188 human serum samples from patients with varying degrees of Leptospirosis presentation [13]. Forty-nine of the discovered 191 reactive antigens were reactive for both IgM and IgG antibodies, and the recognition of these antigens provides a large amount of insight into the mechanism of immune response against leptospiosis [13]. Understanding of this immune response could lead to a better approach to developing an effective vaccine [13]. Further sequencing and analysis of serovar genomes has aided in the development of a greater understanding of Leptospirosis caused by L. interrogans, but there is still much to be discovered regarding this pathogen, and researchers around the world are working diligently to add to the literature surrounding it so that we may more readily and effectively prevent and treat Leptospirosis.
Further Reading
[1]—Genomic Center for Infectious Diseases, Leptospira Genomics and Human Health
References
1. "Leptospirosis Signs and Symptoms." Centers for Disease Control and Prevention. Centers for Disease Control and Prevention, 17 June 2011. Web. 21 Mar. 2015.
2. "VetBact." VetBact. N.p., n.d. Web. 22 Mar. 2015.
3. "Leptospirosis in Humans." VetMed. N.p., n.d. Web. 22 Mar. 2015.
4. Nascimento, A. L. T. O. et al. “Comparative Genomics of Two Leptospira Interrogans Serovars Reveals Novel Insights into Physiology and Pathogenesis .” Journal of Bacteriology 186.7 (2004): 2164–2172. PMC. Web. 26 Feb. 2015.
5. Arean, Victor M., M.D. "The Pathologic Anatomy and Pathogenesis of Fatal Human Leptospirosis (Weil's Disease)." The American Journal of Pathology 40.4 (1962): 393- 423. Web. 25 Feb. 2015.
6. Gamberini, Marcia, Ricardo M. Gomez, Marina V. Atzingen, Elizabeth AL Martins, Silvio A. Vasconcellos, Eliete C. Romero, Luciana C.C. Leite, Paulo L. Ho, and Ana L.T.O Nascimento. "Whole-genome Analysis of Leptospira Interrogans to Identify Potential Vaccine Candidates against Leptospirosis." FEMS Microbiology Letters 244.2 (2005): 305-13. Web. 24 Feb. 2015.
7. Bharti, Ajay R., Jarlath E. Nally, Jessica N. Ricaldi, Michael A. Matthias, Monica M. Diaz, Michael A. Lovett, Paul N. Levett, Robert H. Gilman, Michael R. Willig, Eduardo Gotuzzo, and Joseph M. Vinetz. "Leptospirosis: A Zoonotic Disease of Global Importance." The Lancet Infectious Diseases 3.12 (2003): 757-71. Web. 25 Feb. 2015.
8. Ren, Shuang-Xi, Gang Fu, Xiu-Gao Jiang, Rong Zeng, You-Gang Miao, Hai Xu, Yi-Xuan Zhang, Hui Xiong, Gang Lu, Ling-Feng Lu, Hong-Quan Jiang, Jia Jia, Yue-Feng Tu, Ju-Xing Jiang, Wen-Yi Gu, Yue-Qing Zhang, Zhen Cai, Hai-Hui Sheng, Hai-Feng Yin, Yi Zhang, Gen-Feng Zhu, Ma Wan, Hong-Lei Huang, Zhen Qian, Sheng-Yue Wang, Wei Ma, Zhi-Jian Yao, Yan Shen, Bo-Qin Qiang, Qi-Chang Xia, Xiao-Kui Guo, Antoine Danchin, Isabelle Saint Girons, Ronald L. Somerville, Yu-Mei Wen, Man-Hua Shi, Zhu Chen, Jian-Guo Xu, and Guo-Ping Zhao. "Unique Physiological and Pathogenic Features of Leptospira Interrogans Revealed by Whole-genome Sequencing." Nature 422.6934 (2003): 888-93. Nature. Web. 23 Mar. 2015.
9. Davol, Pamela A. "Canine Leptospirosis." Wing-N-Wave Labradors. N.p., n.d. Web. 24 Mar. 2015.
10. Birnbaum, N., S. C. Barr, S. A. Center, T. Schermerhorn, J. F. Randolph, and K. W. Simpson. "Naturally Acquired Leptospirosis in 36 Dogs: Serological and Clinicopathological Features." Journal of Small Animal Practice 39.5 (1998): 231-36. Web. 24 Feb. 2015.
11. Gonçalves-De-Albuquerque, C. F., P. Burth, A. R. Silva, M. Younes-Ibrahim, H. C. Castro-Faria-Neto, and M. V. Castro-Faria. "Leptospira and Inflammation." Mediators of Inflammation: 1-11. Web. 11 April 2015.
12. Toma, Claudia, Nobuhiko Okura, Chitoshi Takayama, and Toshihiko Suzuki. "Characteristic Features of Intracellular Pathogenic Leptospira in Infected Murine Macrophages." Cellular Microbiology: 1783-792. Web. 11 April 2015.
13. Lessa-Aquino, Carolina, Elsio A. Wunder, Janet C. Lindow, Camila B. Rodrigues, Jozelyn Pablo, Rie Nakajima, Algis Jasinskas, Li Liang, Mitermayer G. Reis, Albert I. Ko, Marco A. Medeiros, and Philip L. Felgner. "Proteomic Features Predict Seroreactivity against Leptospiral Antigens in Leptospirosis Patients." Journal of Proteome Research (2014). American Chemical Society. Web. 10 Apr. 2015.
14. Mudadu, Mauricio De Alvarenga, Viviane Carvalho, and Sophie Y. Leclercq. "Nonclassically Secreted Proteins as Possible Antigens for Vaccine Development: A Reverse Vaccinology Approach." Applied Biochemistry and Biotechnology (2015): n. pag. 12 Feb. 2015. Web. 11 Apr. 2015.
Edited by Emily Gratke, a student of Nora Sullivan in BIOL168L (Microbiology) in The Keck Science Department of the Claremont Colleges Spring 2015.
