Pelagibacterales (SAR11): Difference between revisions
No edit summary |
|||
| (12 intermediate revisions by one other user not shown) | |||
| Line 15: | Line 15: | ||
==Description and Significance== | ==Description and Significance== | ||
Pelagibacterales (SAR11) is an order in the Alphaproteobacteria composed of free-living planktonic oligotrophic facultative photochemotroph bacteria. (4)(5) They are most abundant group of planktonic cells in marine systems and possibly the most numerous bacterium in the world. Typically accounting for ~25% of prokaryotic cells in seawater worldwide.(3) | Pelagibacterales (SAR11) is an order in the Alphaproteobacteria composed of free-living planktonic oligotrophic facultative photochemotroph bacteria. (4)(5) They are most abundant group of planktonic cells in marine systems and possibly the most numerous bacterium in the world. Typically accounting for ~25% of prokaryotic cells in seawater worldwide, SAR11 bacteria play an important role in the global carbon cycle.(3) SAR11 bacteria oxidize organic compounds from primary production into CO2. | ||
The order was originally named SAR11 following its discovery in the Sargasso Sea in 1990 by Professor Stephen Giovannoni and colleagues, from Oregon State University.(2)(5) It was first placed in the order of Rickettsiales, but after rRNA gene-based phyogenetic analysis, in 2013 it was raised to the rank of order, and then placed as sister order to the Rickettsiales in the subclass Rickettsidae.(5) | The order was originally named SAR11 following its discovery in the Sargasso Sea in 1990 by Professor Stephen Giovannoni and colleagues, from Oregon State University.(2)(5) It was first placed in the order of Rickettsiales, but after rRNA gene-based phyogenetic analysis, in 2013 it was raised to the rank of order, and then placed as sister order to the Rickettsiales in the subclass Rickettsidae. They are most closely related to the their sister order, the Rickettsiales. (5) | ||
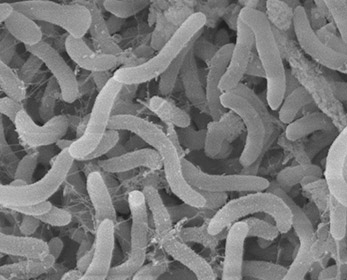
These gram negative, rod shaped bacteria are one of the smallest free cell living organisms, with a cell volume less than 1/500th the volume of E.coli. They have a high surface to volume ratio to better absorb nutrients from its oligotrophic environment. Unsurprisingly, it is also known to have one of the smallest genomes of free living cells.(7) | These gram negative, rod shaped bacteria are one of the smallest free cell living organisms (<0.7 um), with a cell volume less than 1/500th the volume of E.coli. They have a high surface to volume ratio to better absorb nutrients from its oligotrophic environment. These organisms are difficult to grow in pure culture due to their environmental adaptation to such low nutrients and slow growth. Unsurprisingly, it is also known to have one of the smallest genomes of free living cells.(7) The genome of the SAR11 clade has been generated by the DOE Joint Genome Institute. It was found that the isolates formed three micro clusters, closely related groupings within the clade. They were surprised to find that unlike marine SAR11 bacteria, the freshwater bacteria were much less genetically diverse, when measured based on the ratio of recombination to mutation in their genes.(9) It was also found that the bacteria have the complete biosynthetic pathway for all 20 amino acids and all but a few cofactors. There are also no pseudogenes, introns, transposons, or extrachromosomal elements yet observed for any cell. It is the most genetically efficient order of bacteria. | ||
==Subgroups== | ==Subgroups== | ||
Currently the SAR11 clade is divided into five subgroups ( | Currently the SAR11 clade is divided into five subgroups (8): | ||
Subgroup Ia: HTCC1062, HTCC1002, HTCC9565, HTCC7211, HIMB5 | Subgroup Ia: HTCC1062, HTCC1002, HTCC9565, HTCC7211, HIMB5 | ||
Subgroup Ib: SAR193, SAR11 | |||
Subgroup II: Artic95B-1, SAR11 | Subgroup II: Artic95B-1, SAR11 | ||
Subgroup IIIa: HIMB114, OM155 | Subgroup IIIa: HIMB114, OM155 | ||
Subgroup IIIb: S9D-28, LD12 | |||
Subgroup IV: DQ009255 | Subgroup IV: DQ009255 | ||
| Line 36: | Line 38: | ||
Subgroup V: HIMB59, DQ009262 | Subgroup V: HIMB59, DQ009262 | ||
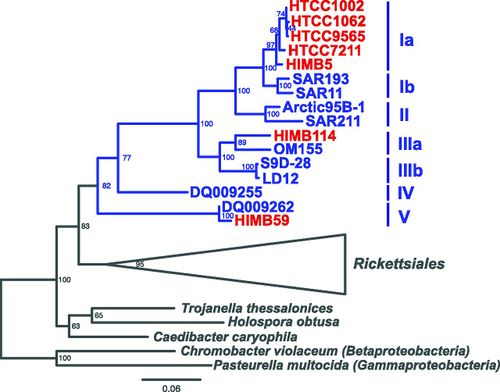
[[Image:SAR11Phylo.jpg|thumb|500px|FIGURE 3: 16S phylogenetic tree of the SAR11 clade (blue). SAR11 has three divergent phylogenetic lineages of the proposed family “Pelagibacteraceae”. Within the SAR11 clade there are 5 subgroups ( | [[Image:SAR11Phylo.jpg|thumb|500px|FIGURE 3: 16S phylogenetic tree of the SAR11 clade (blue). SAR11 has three divergent phylogenetic lineages of the proposed family “Pelagibacteraceae”. Within the SAR11 clade there are 5 subgroups (8).]] | ||
SAR11 clade possesses many unusual features for a free-living organism, including an extremely small, streamlined genome with few paralogs, no pseudogenes, and many missing genes and pathways that are otherwise common in bacteria(8). However, the SAR11 clade is phylogenetically diverse, spanning 18% 16S rRNA gene divergence. They have greater genomic rearrangement at operon boundaries than within operons, as well as hyper variable regions, possibly allowing these organisms to acquire new genetic material with adaptive significance (8). | |||
==References== | ==References== | ||
| Line 68: | Line 56: | ||
(7) [http://www.sciencemag.org.ezproxy.library.wisc.edu/content/309/5738/1242.long Kakarova, K., et al. “Genome Streamlining in a Cosmopolitan Oceanic Bacterium. Stephen J. Giovannoni1,*, H. James Tripp1, Scott Givan2, Mircea Podar3, Kevin L. Vergin1, Damon Baptista3, Lisa Bibbs3, Jonathan Eads3, Toby H. Richardson3, Michiel Noordewier3, Michael S. Rappé4, Jay M. Short3, James C. Carrington2, Eric J. Mathur3. Science 19 August 2005: Vol. 309 no. 5738 pp. 1242-1245 DOI: 10.1126/science.1114057] | (7) [http://www.sciencemag.org.ezproxy.library.wisc.edu/content/309/5738/1242.long Kakarova, K., et al. “Genome Streamlining in a Cosmopolitan Oceanic Bacterium. Stephen J. Giovannoni1,*, H. James Tripp1, Scott Givan2, Mircea Podar3, Kevin L. Vergin1, Damon Baptista3, Lisa Bibbs3, Jonathan Eads3, Toby H. Richardson3, Michiel Noordewier3, Michael S. Rappé4, Jay M. Short3, James C. Carrington2, Eric J. Mathur3. Science 19 August 2005: Vol. 309 no. 5738 pp. 1242-1245 DOI: 10.1126/science.1114057] | ||
(8) [http://mbio.asm.org/content/3/5/e00252-12.full Grote J, et al. 2012. Streamlining and core genome conservation among highly divergent members of the SAR11 clade. mBio 3(5):e00252-12. doi:10.1128/mBio.00252-12.] | |||
(9) [http://jgi.doe.gov/fresh-water-marine-sar11-bacteria-distant-relatives-different-lives Zaremba-Niedzwiedzka K et al. Single-cell genomics reveal low recombination frequencies in freshwater bacteria of the SAR11 clade. Genome Biology 2013, 14:R130 | |||
doi: 10.1186/gb-2013-14-11-r130] | |||
==Author== | ==Author== | ||
Page authored by Digvinder Singh Mavi student of Prof. Katherine Mcmahon at University of Wisconsin-Madison. | Page authored by Digvinder Singh Mavi student of Prof. Katherine Mcmahon at University of Wisconsin-Madison. | ||
<!-- Do not remove this line-->[[Category:Pages edited by students of Katherine | <!-- Do not remove this line-->[[Category:Pages edited by students of Katherine McMahon at University of Wisconsin-Madison]] | ||
Latest revision as of 19:54, 15 October 2015
Classification
Domain: Bacteria
Phylum: Proteobacteria
Class: Alphaproteobacteria
Subclass: Rickettsidae
Order: Pelagibacterales
Description and Significance
Pelagibacterales (SAR11) is an order in the Alphaproteobacteria composed of free-living planktonic oligotrophic facultative photochemotroph bacteria. (4)(5) They are most abundant group of planktonic cells in marine systems and possibly the most numerous bacterium in the world. Typically accounting for ~25% of prokaryotic cells in seawater worldwide, SAR11 bacteria play an important role in the global carbon cycle.(3) SAR11 bacteria oxidize organic compounds from primary production into CO2.
The order was originally named SAR11 following its discovery in the Sargasso Sea in 1990 by Professor Stephen Giovannoni and colleagues, from Oregon State University.(2)(5) It was first placed in the order of Rickettsiales, but after rRNA gene-based phyogenetic analysis, in 2013 it was raised to the rank of order, and then placed as sister order to the Rickettsiales in the subclass Rickettsidae. They are most closely related to the their sister order, the Rickettsiales. (5)
These gram negative, rod shaped bacteria are one of the smallest free cell living organisms (<0.7 um), with a cell volume less than 1/500th the volume of E.coli. They have a high surface to volume ratio to better absorb nutrients from its oligotrophic environment. These organisms are difficult to grow in pure culture due to their environmental adaptation to such low nutrients and slow growth. Unsurprisingly, it is also known to have one of the smallest genomes of free living cells.(7) The genome of the SAR11 clade has been generated by the DOE Joint Genome Institute. It was found that the isolates formed three micro clusters, closely related groupings within the clade. They were surprised to find that unlike marine SAR11 bacteria, the freshwater bacteria were much less genetically diverse, when measured based on the ratio of recombination to mutation in their genes.(9) It was also found that the bacteria have the complete biosynthetic pathway for all 20 amino acids and all but a few cofactors. There are also no pseudogenes, introns, transposons, or extrachromosomal elements yet observed for any cell. It is the most genetically efficient order of bacteria.
Subgroups
Currently the SAR11 clade is divided into five subgroups (8):
Subgroup Ia: HTCC1062, HTCC1002, HTCC9565, HTCC7211, HIMB5
Subgroup Ib: SAR193, SAR11
Subgroup II: Artic95B-1, SAR11
Subgroup IIIa: HIMB114, OM155
Subgroup IIIb: S9D-28, LD12
Subgroup IV: DQ009255
Subgroup V: HIMB59, DQ009262
SAR11 clade possesses many unusual features for a free-living organism, including an extremely small, streamlined genome with few paralogs, no pseudogenes, and many missing genes and pathways that are otherwise common in bacteria(8). However, the SAR11 clade is phylogenetically diverse, spanning 18% 16S rRNA gene divergence. They have greater genomic rearrangement at operon boundaries than within operons, as well as hyper variable regions, possibly allowing these organisms to acquire new genetic material with adaptive significance (8).
References
(1) "Rebounding bacteria". 2013
(2) “SAR11 bacteria thrive — despite viruses”.“Nature”
(6) A war without end - with Earth’s carbon cycle held in the balance
(9) [http://jgi.doe.gov/fresh-water-marine-sar11-bacteria-distant-relatives-different-lives Zaremba-Niedzwiedzka K et al. Single-cell genomics reveal low recombination frequencies in freshwater bacteria of the SAR11 clade. Genome Biology 2013, 14:R130 doi: 10.1186/gb-2013-14-11-r130]
Author
Page authored by Digvinder Singh Mavi student of Prof. Katherine Mcmahon at University of Wisconsin-Madison.