User:S4188552: Difference between revisions
| Line 71: | Line 71: | ||
8. [http://cid.oxfordjournals.org/content/31/3/839.long Bhatti MA, Frank MO (2000) Veillonella parvula meningitis: case report and review of Veillonella infections. Clin Infect Dis. <b>31</b>(3):839-40.] | 8. [http://cid.oxfordjournals.org/content/31/3/839.long Bhatti MA, Frank MO (2000) Veillonella parvula meningitis: case report and review of Veillonella infections. Clin Infect Dis. <b>31</b>(3):839-40.] | ||
10. [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4374535/ Thuy Do, Sheehy EC, Tonnie M, Hughes F, Beighton D (2015) Transcriptomic analysis of three Veillonella spp. present in carious dentine and in the saliva of caries-free individuals. Front Cell Infect Microbiol. <b>5</b:25.] | |||
<references/> | <references/> | ||
This page is written by Aimee Davidson for the MICR3004 course, Semester 2, 2016 | This page is written by Aimee Davidson for the MICR3004 course, Semester 2, 2016 | ||
Revision as of 00:10, 21 September 2016
Aimee Davidson Bench E Date [1]
Classification
Higher order taxa
Kingdom - Bacteria - Firmicutes - Negativicutes - Selenomonadales - Veillonellaceae - Veillonella [3]
Species
Species name: Veillonella parvula (subspecies parvula) [3]
Type strain: ATCC 10790 = CCUG 5123 = DSM 2008 = JCM 12972 = NCTC 11810 [3]
Description and significance
Give a general description of the species (e.g. where/when was it first discovered, where is it commonly found, has it been cultured, functional role, type of bacterium [Gram+/-], morphology, etc.) and explain why it is important to study this microorganism. Examples of citations [1], [2]
Genome structure
Select a strain for which genome information (e.g. size, plasmids, distinct genes, etc.) is available.
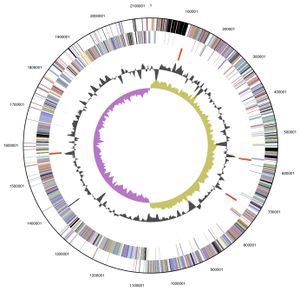
V. parvula exhibits a circular 2.13 Mb (2,132,142 bp) genome, of which 88.46 % is DNA encoding, and was the first member of the Veillonella family to have its complete genome sequenced. Its 1920 genes include: 1859 protein-encoding genes, 61 RNA specific genes and 15 pseduogenes, with an overall GC content of 38.63%. The majority of these genes are predicted to have either an unknown (385 genes) or general function (177 genes). Although those genes with a predicted function are predominately involved with: translation, ribosomal structure, cell wall or membrane biogenesis, energy production and conversion, amino acid, coenzyme and inorganic transport and metabolism [5].
Cell structure and metabolism
Cell wall, biofilm formation, motility, metabolic functions.
Ecology
Aerobe/anaerobe, habitat (location in the oral cavity, potential other environments) and microbe/host interactions.
Pathology
Do these microorganisms cause disease in the oral cavity or elsewhere?
V. parvula has a well established association with human oral diseases, in particular demonstrating a strong association with severe early childhood carries and intraradicular infections [5],[6].
Although V. parvula is generally presented as a commensal or non-pathogenic resident in the bacterial communities of the oropharynx and the gastrointestinal, genitourinary and respiratory tracks [7] , it has rarely been associated with a number of serious and life-threatening infections. These infections include: osteomyeltitis, spondylodiscitis, discitis, meningitis and sinus infections [7], [8].
Application to biotechnology
Bioengineering, biotechnologically relevant enzyme/compound production, drug targets,…
Current research
Summarise some of the most recent discoveries regarding this species.
References
References examples
3. List of prokaryotic names with standing in nomenclature
- ↑ MICR3004
This page is written by Aimee Davidson for the MICR3004 course, Semester 2, 2016
