User:S4188552: Difference between revisions
| Line 14: | Line 14: | ||
'''Species name''': ''Veillonella parvula'' (subspecies ''parvula'') <sup>[[#References|[3]]]</sup> | '''Species name''': ''Veillonella parvula'' (subspecies ''parvula'') <sup>[[#References|[3]]]</sup> | ||
'''Type strain''': ATCC 10790 = CCUG 5123 = DSM 2008 = JCM 12972 = NCTC 11810 <sup>[[#References|[3]]]</sup> | '''Type strain''': Te3 = ATCC 10790 = CCUG 5123 = DSM 2008 = JCM 12972 = NCTC 11810 <sup>[[#References|[3]]]</sup> | ||
==Description and significance== | ==Description and significance== | ||
Revision as of 01:15, 22 September 2016
Aimee Davidson Bench E 23 Sept 2016 [1]
Classification
Higher order taxa
Bacteria - Firmicutes - Negativicutes - Selenomonadales - Veillonellaceae - Veillonella [3]
Species
Species name: Veillonella parvula (subspecies parvula) [3]
Type strain: Te3 = ATCC 10790 = CCUG 5123 = DSM 2008 = JCM 12972 = NCTC 11810 [3]
Description and significance
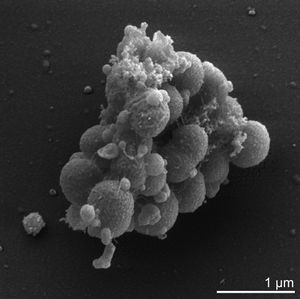
Veillonella parvula is one of thirteen species of the Veillonella genus and one of only six of these species to be found within the human oral cavity[6]. It was original dubbed Staphylococcus parvulus by Veillon and Zuber following its first isolation from an infected human appendix in 1898[11]. It was subsequently renamed Veillonella parvula and further described in 1933 by Prevot[11]. It is a small gram-negative cocci, typically between 0.3 to 0.5 microns in diameter, that can be identified by its characteristic pair grouping and distinctive red fluorescence under UV light and certain growth conditions[4]. As mentioned previously it is commonly found within the human oral cavity, specifically on the tongue, buccal mucosa and in the saliva, however it can also be found within the human gastrointestinal, genitourinary and respiratory tracks[5],[7].
Genome structure
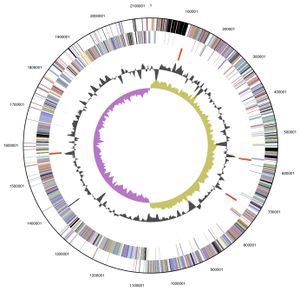
V. parvula exhibits a circular 2.13 Mb (2,132,142 bp) genome, of which 88.46 % is DNA encoding, and was the first member of the Veillonella family to have its complete genome sequenced. Its 1920 genes include: 1859 protein-encoding genes, 61 RNA specific genes and 15 pseduogenes, with an overall GC content of 38.63%. The majority of these genes are predicted to have either an unknown (385 genes) or general function (177 genes). Although those genes with a predicted function are predominately involved with: translation, ribosomal structure, cell wall or membrane biogenesis, energy production and conversion, amino acid, coenzyme and inorganic transport and metabolism [5].
Cell structure and metabolism
Verkley et al established that 98 % of the phosphoglycerides that comprise V. parvula's cell membrane are either serine or ethanolamine based. These phosphoglycerides typically exist in single bonded carbon chains consisting of between 11 to 19 carbons, although chains containing 15,17,18 or 19 carbons atoms may also contain one double carbon bond [12]. Additionally many of these phosphoglycerides exist in a plasmogen form which is thought to have an important role in the regulation of the cell's membrane fluidity[8],[12]. The peptidoglycan cell wall of V. parvula consists of: glutamic acid in the D configuration; diaminopimelic acid in the meso configuration; and alpha-linked cadaverine and putrescine to glutamic acid[5].
Ecology
It is a strict anaerobe whose metabolic activity is centred upon the activity of the methylmalonyl-COA decarboxylase enzyme.
Pathology
V. parvula has a well established association with human oral diseases, in particular demonstrating a strong association with severe early childhood carries and intraradicular infections [5],[6]. Recent research has also revealed that V. parvula is the predominant Veillonella species within dentine biofilms[10]. Although V. parvula generally presents as a commensal or non-pathogenic resident in the bacterial communities of the oropharynx and the gastrointestinal, genitourinary and respiratory tracks [7], it has rarely been associated with a number of serious and life-threatening infections. These infections include: osteomyeltitis, spondylodiscitis, discitis, meningitis and sinus infections [7], [8]. It has demonstrated resistance against a wide range of antibiotics including: tetracycline, erythromycin, gentamicin, karamycin, chloramphenicol and lincomycin. Although importantly at the time of its genome sequencing in 2010 it was still susceptible to penicillin, cephalotin and clindamycin [5].
Application to biotechnology
Bioengineering, biotechnologically relevant enzyme/compound production, drug targets,…
Current research
Summarise some of the most recent discoveries regarding this species.
References
References examples
3. List of prokaryotic names with standing in nomenclature
- ↑ MICR3004
This page is written by Aimee Davidson for the MICR3004 course, Semester 2, 2016
