User:S4352277: Difference between revisions
No edit summary |
No edit summary |
||
| (5 intermediate revisions by the same user not shown) | |||
| Line 11: | Line 11: | ||
===Species=== | ===Species=== | ||
[[Image:K_oralis_gramstain.png|frame|right| | [[Image:K_oralis_gramstain.png|frame|right|150px|Gram stain of <i>Kingella oralis</i> isolated from oral cavity of infected cat.<sup>[[#References|[9]]]</sup>]] | ||
<i>Kingella oralis</i> | <i>Kingella oralis</i> | ||
| Line 25: | Line 25: | ||
==Cell structure and metabolism== | ==Cell structure and metabolism== | ||
The cells of <i>K. oralis</i> are depicted as non-motile agar-corroding rods in sizes of 0.6-0.7 µm x 1-3 µm with rounded ends that can occur in chains or pairs.<sup>[[#References|[2]]]</sup> Although considered non-motile due to the lack of flagella, cells have monopolar fimbriae which | The cells of <i>K. oralis</i> are depicted as non-motile agar-corroding rods in sizes of 0.6-0.7 µm x 1-3 µm with rounded ends that can occur in chains or pairs.<sup>[[#References|[2]]]</sup> Although considered non-motile due to the lack of flagella, cells have monopolar fimbriae which allow for twitching motility.<sup>[[#References|[1]]]</sup> As a gram negative bacterium, <i>K. oralis</i> comprises of the typical structural compositions (lipopolysaccharides, porins, lipoproteins and peptidoglycans). | ||
Typically, members of the <i>Kingella</i> genus ferment glucose and are nutritionally demanding, oxidase-positive and catalase-negative bacterium. <i>Kingella</i> cells produce acid from glucose and other carbohydrates. <sup>[[#References|[5]]]</sup> Unlike <i>Kingella Kingae</i>, <i>K. oralis</i> cannot produce acid from maltose or reduce nitrite and nitrate in its metabolic process. <sup>[[#References|[2]]]</sup> | Typically, members of the <i>Kingella</i> genus ferment glucose and are nutritionally demanding, oxidase-positive and catalase-negative bacterium. <i>Kingella</i> cells produce acid from glucose and other carbohydrates. <sup>[[#References|[5]]]</sup> Unlike <i>Kingella Kingae</i>, <i>K. oralis</i> cannot produce acid from maltose or reduce nitrite and nitrate in its metabolic process. <sup>[[#References|[2]]]</sup> | ||
| Line 35: | Line 35: | ||
<i>K. oralis</i> is a pathogenic microorganism as it has been indicated to be associated with severe periodontitis in humans. As the dominant species of the subgingival microbiota in periodontitis sites, the capacity to cause disease is highlighted. <sup>[[#References|[3]]]</sup> Furthermore, a case study in 2010 has indicated that it has the capacity to infect felines and perhaps other animals. <sup>[[#References|[9]]]</sup> | <i>K. oralis</i> is a pathogenic microorganism as it has been indicated to be associated with severe periodontitis in humans. As the dominant species of the subgingival microbiota in periodontitis sites, the capacity to cause disease is highlighted. <sup>[[#References|[3]]]</sup> Furthermore, a case study in 2010 has indicated that it has the capacity to infect felines and perhaps other animals. <sup>[[#References|[9]]]</sup> | ||
Species belonging to the <i>Kingella</i> genus express multiple virulence factors. A type IV pili is used for enhancing adhesion to epithelial and synovial cells for colonisation. <i>K. oralis</i> forms spreading/corroding colonies with high density pilation by regulating a major pilus subunit, pilA1 through expressing high levels of σ54. Although it may have a direct link to periodontitis, it is less virulent than <i>K. kingae</i> which produces RTX Toxin, a potent cytotoxin. <sup>[[#References|[7]]]</sup> | Species belonging to the <i>Kingella</i> genus express multiple virulence factors. A type IV pili is used for enhancing adhesion to epithelial and synovial cells for colonisation. <i>K. oralis</i> forms spreading/corroding colonies with high density pilation by regulating a major pilus subunit, pilA1 through expressing high levels of σ54.<sup>[[#References|[8]]]</sup> Although it may have a direct link to periodontitis, it is less virulent than <i>K. kingae</i> which produces RTX Toxin, a potent cytotoxin that destroys targeted tissues such as synovium to breach the epithelial barrier. <sup>[[#References|[7]]]</sup> | ||
==Applications to biotechnology== | |||
The utilisation of <i>K. oralis</i> to determine and verify bacterial interactions such as coadhesion and coaggregation of co-localised adhesins and complementary receptors have been mentioned. Apart from this, no other studies have implied further use of the organism in the biotechnology field.<sup>[[#References|[6]]]</sup> | |||
==Current research== | |||
A study conducted in 2013 indicated the presence of a novel dialect of DNA Uptake Sequence (kingDUS) and how it is integral for the down regulation of recombination and DNA uptake in transformation. It was determined that the genome consisted of approximately 2.5% of kingDUS while <i>Neisseriaceae meningitis</i> comprised of only 1% DUS.<sup>[[#References|[10]]]</sup> | |||
Most recent research in 2014 involved the investigation of <i>K. oralis</i> involvement as a coadhesion partner in microbial biofilm communities. It was found that complex biofilms were formed via interbacterial adhesion of <i>K. oralis</i> and <i>Streptococcus</i> bacteria.<sup>[[#References|[6]]]</sup> | |||
==References== | ==References== | ||
1.[https://books.google.com.au/books?id=VloMBAAAQBAJ&pg=PA374&lpg=PA374&dq=kingella+oralis+pathogen&source=bl&ots=6d1gdrdRXo&sig=pO3I-5S_YPZfwJvXReDmfkf2lbo&hl=en&sa=X&ved=0ahUKEwi5y-jWrp3PAhUEOj4KHUt2AWYQ6AEIRjAF#v=onepage&q=kingella%20oralis&f=false Mahon, C.R., Lehman, D.C., Jr, G.M., 2014. Textbook of Diagnostic Microbiology. Elsevier Health Sciences.] | 1.[https://books.google.com.au/books?id=VloMBAAAQBAJ&pg=PA374&lpg=PA374&dq=kingella+oralis+pathogen&source=bl&ots=6d1gdrdRXo&sig=pO3I-5S_YPZfwJvXReDmfkf2lbo&hl=en&sa=X&ved=0ahUKEwi5y-jWrp3PAhUEOj4KHUt2AWYQ6AEIRjAF#v=onepage&q=kingella%20oralis&f=false Mahon, C.R., Lehman, D.C., Jr, G.M., 2014. Textbook of Diagnostic Microbiology. Elsevier Health Sciences.] | ||
| Line 55: | Line 60: | ||
9.[http://www.vetpol.org.pl/prawo-projekty-legislacja/doc_download/727-04-artykul Adaszek, Ł., et al. 2010. A case of Kingella oralis infection in the cat. Życie Weterynaryjne <b>85</b>(7): 604-606] | 9.[http://www.vetpol.org.pl/prawo-projekty-legislacja/doc_download/727-04-artykul Adaszek, Ł., et al. 2010. A case of Kingella oralis infection in the cat. Życie Weterynaryjne <b>85</b>(7): 604-606] | ||
10.[http://journals.plos.org/plosgenetics/article?id=10.1371%2Fjournal.pgen.1003458 Frye, S.A., Nilsen, M., Tønjum, T., Ambur, O.H., 2013. Dialects of the DNA Uptake Sequence in Neisseriaceae. PLOS Genet 9, e1003458.] | |||
<references/> | <references/> | ||
This page is written by Kristoffer Hua for the MICR3004 course, Semester 2, 2016 | This page is written by Kristoffer Hua for the MICR3004 course, Semester 2, 2016 | ||
Latest revision as of 06:03, 23 September 2016
Kristoffer Hua
Bench C
21/09/2016 [1]
Classification
Higher order taxa
Bacteria – Proteobacteria – Betaproteobacteria – Neisseriales– Neisseriaceae – Kingella
Species
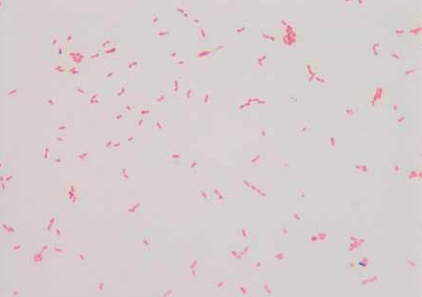
Kingella oralis
Type strain: UB-38, ATCC 51147, CCUG 30450, CIP 103803.
Description and significance
Kingella oralis is a gram negative, bacilli-shaped facultative anaerobe.[1] This organism is found to have high prevalence in human dental plaque, mucosal surfaces and in saliva.[2] Contrary to the other members of the Kingella genus, K. oralis is a common colonizer in the human oral cavity that has little involvement in diseases. Associations of this bacteria and gum diseases such as gingivitis and periodontitis have been recognized. This is inferred because in infection sites, high prevalence is evident. However, correlations to the severity are not clearly distinguished. [3]
Studies in culture indicate that it is a non-motile organism; however the cells are capable of forming spreading colonies on agar.[1] Similarities in phenotypic traits and oral distribution with Eikenella corrodens make it difficult to detect K. oralis with rapid diagnostic assays. [3]
Genome structure
Like most bacteria, K. oralis has a circular genome which contains 2,406,670 base pairs with a GC content of 54.3%. A total of 2371 genes have been recognized, 2315 encoding for proteins and 56 associated with RNA.[4]
Cell structure and metabolism
The cells of K. oralis are depicted as non-motile agar-corroding rods in sizes of 0.6-0.7 µm x 1-3 µm with rounded ends that can occur in chains or pairs.[2] Although considered non-motile due to the lack of flagella, cells have monopolar fimbriae which allow for twitching motility.[1] As a gram negative bacterium, K. oralis comprises of the typical structural compositions (lipopolysaccharides, porins, lipoproteins and peptidoglycans).
Typically, members of the Kingella genus ferment glucose and are nutritionally demanding, oxidase-positive and catalase-negative bacterium. Kingella cells produce acid from glucose and other carbohydrates. [5] Unlike Kingella Kingae, K. oralis cannot produce acid from maltose or reduce nitrite and nitrate in its metabolic process. [2]
Ecology
Colonisation of the oral cavity and the upper respiratory tract and its survivability is determined by a few factors. As a mesophile, the organism can persist in temperatures between 20°C and 45°C.[4] Interbacterial adhesion between genetically distinct bacteria such as streptococci and actinomyces species allow for efficient usage of nutrients and protection. This interaction is instigated by the co-localisation of bacterial adhesins and complementary receptors. [6]
Pathology
K. oralis is a pathogenic microorganism as it has been indicated to be associated with severe periodontitis in humans. As the dominant species of the subgingival microbiota in periodontitis sites, the capacity to cause disease is highlighted. [3] Furthermore, a case study in 2010 has indicated that it has the capacity to infect felines and perhaps other animals. [9]
Species belonging to the Kingella genus express multiple virulence factors. A type IV pili is used for enhancing adhesion to epithelial and synovial cells for colonisation. K. oralis forms spreading/corroding colonies with high density pilation by regulating a major pilus subunit, pilA1 through expressing high levels of σ54.[8] Although it may have a direct link to periodontitis, it is less virulent than K. kingae which produces RTX Toxin, a potent cytotoxin that destroys targeted tissues such as synovium to breach the epithelial barrier. [7]
Applications to biotechnology
The utilisation of K. oralis to determine and verify bacterial interactions such as coadhesion and coaggregation of co-localised adhesins and complementary receptors have been mentioned. Apart from this, no other studies have implied further use of the organism in the biotechnology field.[6]
Current research
A study conducted in 2013 indicated the presence of a novel dialect of DNA Uptake Sequence (kingDUS) and how it is integral for the down regulation of recombination and DNA uptake in transformation. It was determined that the genome consisted of approximately 2.5% of kingDUS while Neisseriaceae meningitis comprised of only 1% DUS.[10]
Most recent research in 2014 involved the investigation of K. oralis involvement as a coadhesion partner in microbial biofilm communities. It was found that complex biofilms were formed via interbacterial adhesion of K. oralis and Streptococcus bacteria.[6]
References
2.Liu, D., 2011. Molecular Detection of Human Bacterial Pathogens. CRC Press.
4.National Center for Biotechnology Information, Kingella oralis overview, viewed 19-09-2016
- ↑ MICR3004
This page is written by Kristoffer Hua for the MICR3004 course, Semester 2, 2016