Prochlorococcus: Difference between revisions
Slonczewski (talk | contribs) No edit summary |
Slonczewski (talk | contribs) No edit summary |
||
| (7 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
{{Curated}} | |||
{{Biorealm Genus}} | {{Biorealm Genus}} | ||
[[Image:may30_chisholm1.jpg|thumb|350px|right|''Prochlorococcus marinus'' MIT 9313. Image | |||
[[Image:may30_chisholm1.jpg|thumb|350px|right|''Prochlorococcus marinus'' MIT 9313. | courtesy of [http://www.williams.edu/Biology/Faculty_Staff/cting/cting.shtml Claire S. Ting, Department of Biology, Williams College.]] | ||
courtesy of [http://www.williams.edu/Biology/Faculty_Staff/cting/cting.shtml Claire S. Ting, Department of Biology, Williams College.]] | |||
==Prochlorococcus== | |||
==Classification== | ==Classification== | ||
| Line 14: | Line 17: | ||
''Prochlorococcus marinus MED4, P. marinus MIT9313, P. sp.'' | ''Prochlorococcus marinus MED4, P. marinus MIT9313, P. sp.'' | ||
{| | {|| height="10" bgcolor="#FFDF95" | | ||
| height="10" bgcolor="#FFDF95" | | |||
'''NCBI: [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=1218 Taxonomy] [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=genomeprj&cmd=Search&dopt=DocSum&term=txid1218%5BOrganism:exp%5D Genome]''' | '''NCBI: [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=1218 Taxonomy] [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=genomeprj&cmd=Search&dopt=DocSum&term=txid1218%5BOrganism:exp%5D Genome]''' | ||
|} | |} | ||
Latest revision as of 14:36, 23 January 2017
A Microbial Biorealm page on the genus Prochlorococcus
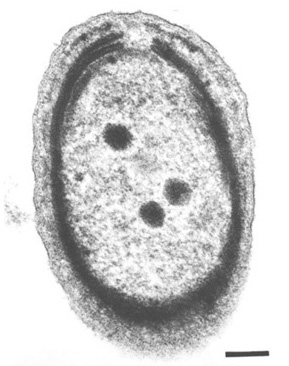
Prochlorococcus
Classification
Higher order taxa:
Bacteria; Cyanobacteria; Prochlorophytes; Prochlorococcaceae; Prochlorococcus
Species:
Prochlorococcus marinus MED4, P. marinus MIT9313, P. sp.
NCBI: Taxonomy GenomeDescription and Significance
Although Prochlorococcus is the smallest known phototroph it contributes 30-80% of primary production in the world's oligotrophic oceans, and is consequently plays a significant role in the global carbon cycle and the Earth's climate.
Genome Structure
The genomes of both Prochlorococcus marinus MED4 and Prochlorococcus marinus MIT9313 have been sequenced, representing both high light-adapted and low light-adapted ecotypes respectively. It appears as though, in prochlorococcus, distribution in the water column is a more important environmental pressure than is horizontal location. Thus researchers have chosen both a high light-adapted organism and a low light-adapted organism for a more specific comparison of the two species. The genome of Prochlorococcus marinus MED4 is approximately 1.67 Mega-base pairs long with 1,694 predicted protein-coding regions. The genome of Prochlorococcus marinus MIT 9313 is about 2.40 Mega-base pairs long, with 2,195 predicted protein-coding regions.
Cell Structure and Metabolism
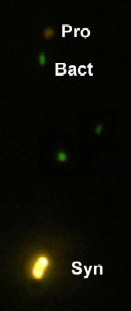
Prochlorophytes are oxygenic photosynthesizers that, along with synechococcus, dominate the photoautotrophic picoplankton in the world's oceans. Synechococcus was discovered first because of its intense orange phycoerythrin fluorescence. Prochlorococcus wasn't discovered until the 1980s when researchers wee able to detect the dim red fluorescence emitted by its unique divinyl derivatives of chlorophyll a and b.
Prochlorophytes have an unorthodox compliment of pigments associated with their photosynthetic apparatus. Instead of the phycobilisomes characteristic of cyanobacteria, they have divinyl derivatives of chlorophyll a and b, a-carotene, zeaxanthin, and a type of phycoerythrin. Thus far, prochlorococcus is the only organism known to contain this particular compliment of pigments. This particular array of pigments enables the phytoplankter to absorb blue light efficiently at low light intensities characteristic of the deep euphotic zone.
Ecology
Prochlorococcus is a tiny, single-celled, cyanobacteria, that inhabits the ocean in a latitudinal band from 40 degrees north to 40 degrees south where they occupy a key position at the base of the food web. But it is thought that they also may help regulate population with their viruses or phages. They are usually absent from brackish or well-mixed waters, and mostly inhabit tropical and subtropical oceans, and extend deep into the water column, often below the oxycline. Prochlorophytes can be found as deep as 150 to 200 meters where only 0.1% of the surface irradiance reaches them.
The important vertical distribution of prochlorococcus from the euphotic zone to below the oxycline has developed different ecotypes uniquely adapted to their position in the water column. In addition to the already unique compliment of pigments adapted to low-light conditions, strains of prochlorococcus adapted to low light, also have the unique ability to accumulate large amounts of chlorophyll b relative to chlorophyll a. These, low-light adapted strains also must cope with low oxygen condition as well. They derive some of their energy from photosynthesis still, but use dissolved organic carbon as a source of reduced carbon and energy.
In accordance to their specific adaptations to their position in the water column is the inability of high-light adapted species to utilize nitrate or nitrite as a nitrogen source. When cultured, high light-adapted species could grow in a medium with ammonium, but could not grow on a medium where nitrate or nitrite were the only nitrogen source. Low light-adapted strains on the other hand, could grow in mediums of nitrite in addition to ammonium and urea. This might reflect the nitrite maximum found at the bottom of the euphotic zone.
References
DOE Joint Genome Institute: Prochlorococcus marinus MED4 Genome Project Completed Genome
DOE Joint Genome Institute: Prochlorococcus marinus MIT 9313 Genome Project Completed Genome
Fuhrman, Jed A. & Douglas Capone. 2001. Nifty nanoplankton. Nature, 412: 593-594.
Genome Project Home Page: Prochlorococcus marinus SS120
Genoscope: Prochlorococcus marinus - complete genome
Goericke, R. et al. 2000. A novel niche for Prochlorococcus sp. in low-light suboxic environments in the Arabian Sea and the Eastern Tropical North Pacific. Deep-Sea Research, 47: 1183-1205.
Karl, David M. 2002. Hidden in a sea of microbes. Nature, 415: 590-591.
