Lactobacillus sanfranciscensis: Difference between revisions
| Line 4: | Line 4: | ||
]] | ]] | ||
<br>This page was curated by <b>Kay Burrows</b> for Dr. Joan Slonczewski's Microbiology Class, Spring 2018.<br> | <br>This page was curated by <b>Kay Burrows</b> for Dr. Joan Slonczewski's Microbiology Class, Spring 2018.<br> | ||
<br>There are over 180 species of <i>Lactobacillus</i> currently documented. <i>Lactobacillus</i> can be found in the human reproductive system as well as the urinary and digestive tracts. A major contributor to gut flora, <i>Lactobacilli</i> have numerous beneficial roles in the human gastrointestinal tract, including immunomodulation and nutrient breakdown. [[#References|[3]]] More than 50 species of <i>Lactobacilli</i> are known as useful non-pathogenic bacteria for the processing of different foods, or for medical application in restoring normal, healthy flora to the skin, vaginal biofilm, and digestive tract.<ref>[https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3417654/pdf/fcimb-02-00086.pdf : Pessione E. Lactic acid bacteria contribution to gut microbiota complexity: lights and shadows. Frontiers in Cellular and Infection Microbiology. 2012]</ref> Many species within Lactobacillus have a high hydrogen peroxide tolerance, lending further to their role in inhibiting potential pathogens. | <br>There are over 180 species of <i>Lactobacillus</i> currently documented. <i>Lactobacillus</i> can be found in the human reproductive system as well as the urinary and digestive tracts. A major contributor to gut flora, <i>Lactobacilli</i> have numerous beneficial roles in the human gastrointestinal tract, including immunomodulation and nutrient breakdown. [[#References|[3]]] More than 50 species of <i>Lactobacilli</i> are known as useful non-pathogenic bacteria for the processing of different foods, or for medical application in restoring normal, healthy flora to the skin, vaginal biofilm, and digestive tract.<ref>[https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3417654/pdf/fcimb-02-00086.pdf : Pessione E. Lactic acid bacteria contribution to gut microbiota complexity: lights and shadows. Frontiers in Cellular and Infection Microbiology. 2012]</ref> Many species within <i>Lactobacillus</i> have a high hydrogen peroxide tolerance, lending further to their role in inhibiting potential pathogens. | ||
Among this genus is a unique and popular bacterium, <i>Lactobacillus sanfranciscensis </i>. <i>L. sanfranciscensis</i> is a rod-shaped, Gram positive, lactic acid bacteria, closely related to the <i>Lactobacillus casei-Pediococcus</i> group. It is nonsporulating, nonrespiring, acid- and aero-tolerant. Famous for being an important flavor component of sourdough bread, <i>L. sanfranciscensis</i> is an obligate, heterofermentative bacteria.[[#References|[6]]] This heterofermentation, or fermentation that produces more than one product, creates both lactic acid and, in certain conditions, ethanol and carbon dioxide. [[#References|[8]]] It makes ample use of resources by utilizing a symbiotic relationship with different yeasts, including <i>Saccharomyces cerevisiae</i> and <i>Candida milleri</i>. This yeast, also useful in the making of sourdough, provides <i>L. sanfranciscensis</i> with amino acids and peptides to aid in growth and fermentation. | Among this genus is a unique and popular bacterium, <i>Lactobacillus sanfranciscensis </i>. <i>L. sanfranciscensis</i> is a rod-shaped, Gram positive, lactic acid bacteria, closely related to the <i>Lactobacillus casei-Pediococcus</i> group. It is nonsporulating, nonrespiring, acid- and aero-tolerant. Famous for being an important flavor component of sourdough bread, <i>L. sanfranciscensis</i> is an obligate, heterofermentative bacteria.[[#References|[6]]] This heterofermentation, or fermentation that produces more than one product, creates both lactic acid and, in certain conditions, ethanol and carbon dioxide. [[#References|[8]]] It makes ample use of resources by utilizing a symbiotic relationship with different yeasts, including <i>Saccharomyces cerevisiae</i> and <i>Candida milleri</i>. This yeast, also useful in the making of sourdough, provides <i>L. sanfranciscensis</i> with amino acids and peptides to aid in growth and fermentation. | ||
Revision as of 00:48, 5 May 2018
Lactobacillus sanfranciscensis : The Key to the Perfect Sourdough

This page was curated by Kay Burrows for Dr. Joan Slonczewski's Microbiology Class, Spring 2018.
There are over 180 species of Lactobacillus currently documented. Lactobacillus can be found in the human reproductive system as well as the urinary and digestive tracts. A major contributor to gut flora, Lactobacilli have numerous beneficial roles in the human gastrointestinal tract, including immunomodulation and nutrient breakdown. [3] More than 50 species of Lactobacilli are known as useful non-pathogenic bacteria for the processing of different foods, or for medical application in restoring normal, healthy flora to the skin, vaginal biofilm, and digestive tract.[2] Many species within Lactobacillus have a high hydrogen peroxide tolerance, lending further to their role in inhibiting potential pathogens.
Among this genus is a unique and popular bacterium, Lactobacillus sanfranciscensis . L. sanfranciscensis is a rod-shaped, Gram positive, lactic acid bacteria, closely related to the Lactobacillus casei-Pediococcus group. It is nonsporulating, nonrespiring, acid- and aero-tolerant. Famous for being an important flavor component of sourdough bread, L. sanfranciscensis is an obligate, heterofermentative bacteria.[6] This heterofermentation, or fermentation that produces more than one product, creates both lactic acid and, in certain conditions, ethanol and carbon dioxide. [8] It makes ample use of resources by utilizing a symbiotic relationship with different yeasts, including Saccharomyces cerevisiae and Candida milleri. This yeast, also useful in the making of sourdough, provides L. sanfranciscensis with amino acids and peptides to aid in growth and fermentation.
During fermentation, the microbe consumes maltose and glucose to ultimately release acids that create the sour taste in sourdough.[3] L. sanfranciscensis, contains a very small genome and a relatively high ratio of rRNA operons, crucial for rapid growth. Unlike the majority of homofermentative Lactobacilli, L. sanfranciscensis uses heterofermentative metabolism, processing sugars into either lactic acid or alcohol. This unique processing is what contributes to its role in sourdough formation. [4]
Ultimately, we will aim to answer the question: How do we form the ideal sourdough using these microbes?
Discovering sanfranciscensis
Though humans have been making bread with flour and water for 5,000 years, the science behind this magical process was uncovered much later. In the 1800s, Louis Pasteur uncovered the importance of yeast and fermentation in the famous bread rise. The most robust of these yeast for this purpose, Saccharomyces cerevisiae, has since been purified and mass produced for use in bread products around the world. Microbes that contribute to the flavor and characteristics of different breads, however, are more mysterious. Different starters can contain a multitude of bacteria and yeast in differing proportions to give breads a recognizable flavor. These components were once reliant on the available microbes in the region in which the bread was produced, but are now available worldwide. Lactobacillus sanfranciscensis is one of such bacteria that became popular for its distinctive tang contribution-- perfect for sourdough bread. In fact, in a sourdough culture, L. sanfranciscensis can outnumber its yeast counterpart 100 to 1.
Sourdough production began as a method for processing cereals. Without the addition of yeast, ancient grains such as spelt and barley created flat, sour breads.Many of these breads are still made and consumed today, such as Injera, the flat, brown bread popular in Ethiopia. Variations grew from this flat bread, using different grains depending on the region. Rye bread, requiring inhibition of a-amylase and pentosans, is still popular in Central Europe as a result of the spread of sourdough. Once brewing became more popular, sour bread combined with wheat flour and yeast to create the leavened sourdough bread we know today. Since sourdough stayed fresh longer than other breads, it was popularized in the long trips of the California Gold Rush. [5]
Originally, it was assumed that the famous L. sanfranciscensis, discovered and popularized in San Francisco, was unique to the Bay Area. San Francisco was producing great numbers of phenomenally complex sourdoughs, and an effort was made by several groups to uncover the composition of their starters. The results were in, and Lactobacillus sanfranciscensis seemed to be the defining microbe of this west coast phenomenon. It was first isolated by Kline and Sugihara in 1971, who characterized and named the bacteria. However, it was later discovered by the Sourdough Project that L. sanfranciscensis could be found in nearly 90 other countries-- either spread from other successful sourdough starters, or native to the regions themselves. L. sanfranciscensis is, in fact, identical to separately-identified European strain Lactobacillus brevis var. lindneri. [6]
The San Francisco name stuck, despite the bacteria’s lack of specificity to that region.
Lactobacillus sanfranciscensis in a classic San Francisco sourdough bread is often used with yeast strain Candida milleri. Since C. milleri is particularly acid tolerant, it can handle increasing lactic acid concentrations and lower pH that result from L. sanfranciscensis growth and metabolism. Furthermore, Candida milleri cannot digest maltose from flour starch. This sugar is luckily necessary for L. sanfranciscensis survival, which makes a pairing of this yeast and bacterium particularly symbiotic. Regardless, most larger bakeries still maintain S. cerevisiae as their primary yeast in sourdough breads, due to its simplicity and availability. Some of such bakeries rely on last minute acetic acid additions to achieve the ultimate tangy taste that would have been produced by a more ideal pairing.
Contribution to Sourdough Starter
In sourdough bread making, the starter determines quite a bit of the bread’s flavor, texture, and aroma. Different starters containing unique combinations of bacteria and yeast are responsible for the many of the differences in bread types. In sourdough bread, as we know, the main flavor contributing microbes are lactic acid bacteria, contributing a sharp sour taste from high acidity. Other contributions to the bread’s texture and flavor include the types of flour used, the conditions of the bread rise, and a slew of additional ingredients optional to the breadmaker.
Among sourdough starters, as little as 30% and as many as 95% of the lactic acid bacteria have been recorded to be Lactobacillus sanfranciscensis. This high percentage of L. sanfranciscensis may be due in part to the species’ antimicrobial activity. Though homofermentative bacteria generally have greater antimicrobial properties against coliforms than heterofermentative bacteria, L. sanfranciscensis produces a bacteriocin-like inhibitory substance (BLIS C57) that has bacteriocidal and bacteriolytic properties. All known lactic acid bacteria strains in sourdough, other than Lactobacillus fructivorans, are sensitive to this compound. Therefore, production of BLIS C57 by L. sanfranciscensis likely contributes to its ability to dominate a sourdough starter. [7]
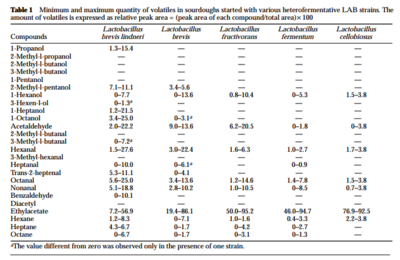
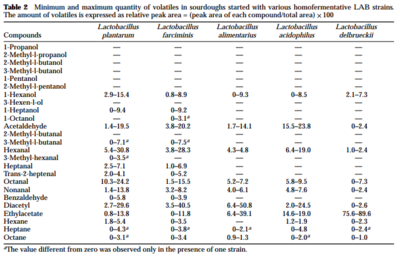
L. sanfranciscensis also contributes to sourdough volatile compounds, important for both flavor and aroma. Though the flavor of the original wheat, temperature during fermentation, and baking contribute the largest amount of aroma to a bread, microbes cannot be underrated. In a study of 87 species of lactic acid bacteria, Gobbetti et al (1995) found that certain types of bacteria contributed different volatile compounds. [8] Without lactic acid bacteria, the breads presented aroma compounds of “acetaldehyde, hexanal, octanal, nonanal and ethylacetate”. These compounds were also found in sourdoughs with lactic acid bacteria, but in significantly higher amounts. Yeast fermentation itself contributes iso-alcohols as well. Table 1 and 2 (right) demonstrate the different compounds contributes by specific species of lactic acid bacteria. In general, increased concentrations of ethylacetate, alcohols, and aldehydes were due to contribution of heterofermentative lactic acid bacteria. On the other hand, homofermentative lactic acid bacteria contributes diacetyl and other carbonyls. The study highlights two specific strains, L.breviss and L. plantarum, as two bacteria with particularly extensive contributions to volatile compound profile. Furthermore, both heterofermentative and homofermentative lactic acid bacteria helped enhance the volatile compounds released by yeast alone. This supports that a starter rich in different varieties of lactic acid bacteria can help contribute unique compounds to the bread’s aroma and flavor.
Sourdough starters are often considered like an heirloom-- bakers will start their own starter as a gift or purchase of a population of microbes from another baker. However, research from Ripari (2016) has been conducted to determine the ability to organically start a sourdough starter from other sources.
[9]The use of fruits, flowers, or other plants have been used to select for certain microorganisms and encourage microbial evolution in sourdough starters. In their study, the group found that plant material frequently resulted in rapidly stable microbial communities in sourdough starters. Furthermore, in four of the seven organically-started inoculates, L. sanfranciscensis quickly dominated the microbial scene. One interesting addition was that sourdoughs started with apple flowers or pulp saw significant contribution from Acetobacter, not found in any other plant inoculates. Others saw higher populations of L. plantarum, graminis, and rossiae, along with the L. sanfranciscensis. S. cerevisiae was organically cultured in every inoculate.[10]
Genome & Metabolism
Lactic acid bacteria were once classified by chemotaxonomical properties including cell wall composition or cellular fatty acid composition. Though phenotype could distinguish between genuses, it did not have very much taxonomic usefulness, as lactic acid bacteria can be very closely related. Thus, DNA-based techniques, such as locating rRNA genes and GC-content analysis, has been necessary to differentiate between different lactic acid bacteria. Furthermore, identification of housekeeping genes has proven useful for classification of lactic acid bacteria. Since over 180 species of Lactobacilli have been identified, these characteristic are important in the classification and naming of different species within this genus. (Huys 2012) [11]
The genome of L. sanfranciscensis was analyzed by Vogel et al (2011). They found that L. sanfranciscensis had a circular chromosomal sequence length of 1,298,316 bp, as well as two plasmids of lengths 58,739 bp and 18,715 bp. This particularly small genome makes it the smallest of such within the Lactobacilli genus. Vogel’s group also discovered that the L. sanfranciscensis genome contained seven rRNA operons and 61 tRNA genes-- the highest density of RNA genes in all sequenced free-living organisms known to date at 5.39 per Mbp. These rRNA operons allow L. sanfranciscensis to grow and adapt rapidly to the sourdough environment. This is further supported by the increased number of genes for oxidative stress response. The species also did not have a high number of genes for regulation, which might imply that they have adapted to a nutrient-rich environment and no longer require strict regulation. [12]
Evidence for heterofermentation as a mechanism of metabolism in L. sanfranciscensis was also found by Vogel’s group. Genes necessary for the phosphoketolase pathway were identified, but there were not genes for transaldolase or transketolase. L. sanfranciscensis can thus use fructose, ribose and maltose for energy. Furthermore, genes for oligo-1,6-glucosidase show that the species can also hydrolyse a-1,6-D-glucosidic bonds found in starch and glycogen. Among lactic acid bacteria, sugar can be transported by primary or secondary transport systems, or phosphoenolpyruvate:sugar phosphate transferase systems. In primary transport systems, ATP is used to translocate sugars across the membrane. By contrast, secondary transport systems drive sugar transport with electrochemical gradients. Phosphoenolpyruvate:sugar phosphotransferase systems which use phosphoenol pyruvate to phosphorylate sugars that can then be translocated. Neubauer et al (1994) found that L. sanfranciscensis demonstrates a secondary transport system-- or more specifically, a maltose-H+ symport and glucose uniport system. [13] The main sugar in sourdough, maltose, is taken up by a constitutively expressed maltose carrier protein. Maltose is then degraded into glucose and glucose 1-phosphate by maltose phosphorylase, which are excreted only up to the equilibration level.
L. sanfranciscensis can also use fructose as an electron acceptor. In this pathway, fructose is reduced to mannitol, and the cell can produce less ethanol through maltose fermentation. With a lower concentration of ethanol, can use the acetate kinase reaction to produce ATP and synthesize acetic acid. A similar pathway can be utilized if the cell has access to oxygen as an electron acceptor. A greater amount of acetic acid can thus be produced if the original mixture has fructose added or is better aerated.
The degradation of these sugars is a crucial aspect of the sourdough process. Without it, the large sugars in the flour would remain undigested, making for further difficulty in human digestion. Furthermore, the yeast would be unable to utilize the maltose without it being further broken down into glucose molecules. The bread would be unable to properly rise without yeast fermentation releasing carbon dioxide bubbles throughout the mixture. Thus, the very essence of bread formation-- particularly the bubbly, easy-to-digest sourdough, relies on L. sanfranciscensis.
Lactic acid production, however, is easily the most important contribution of L. sanfranciscensis. In 1857, Louis Pasteur discovered that lactic acid was a product of microbial fermentation. From there, we grew to understand that bacteria participating in anaerobic fermentation could experience heterofermentative or homofermentative metabolism, as described earlier. In a homofermentative process, a six-carbon sugar is converted into two lactic acid molecules, creating two ATP molecules along the way. In a heterofermentative process, one lactate molecule is produced from one six-carbon sugar, as well as carbon dioxide and either acetic acid or ethanol. These variations in heterofermentation are due to conditions surrounding the bacteria and potential electron donors. At a higher temperature and higher water content, this process leans toward alcohol production. By contrast, at cooler temperatures and drier conditions, heterofermentation is likely to produce more acetic acid. [14] Therefore, L. sanfranciscensis, in its processing of six carbon sugars in an anaerobic environment, produces lactic acid as a waste product, and we use this waste product to flavor delicious varieties of bread!
Growing Conditions
Lactobacillus sanfranciscensis has been cultured for many years through a procedure called “backslopping”-- that is, the repetitive re-inoculation of a starter by adding flour and water, and never using the entire starter at any given time. This ensures that the bacteria are given plenty of nutrients to grow and multiply, prolonging the life of the starter. Over time, backslopping produces a steady ratio of certain bacteria, and thus a baker develops his or her own special flavor. Commericially-sold starters often include the basics, along with L. delbrueckii, fermentum, and plantarum. [15]
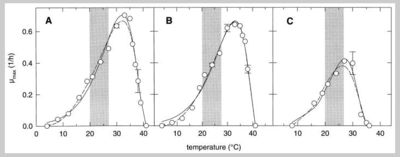
Perfect function of L. sanfranciscensis relies on a balance of certain environmental conditions. Though bakers can specialize in certain techniques, additions, and temperatures, Ganzel et al (1998) revealed that L. sanfranciscensis itself does have an ideal environment to perform at its best, albeit broad. The group studied the contribution of ionic strength, pH, temperature, and metabolic end products (lactate, acetate, and ethanol) on the stability and metabolism of L. sanfranciscensis and its yeast counterpart C. milleri. They found that L. sanfranciscensis grew best between 30 and 37°C, whereas C. milleri responded well to temperatures below 26°C -- this is consistent with the “baker’s rule” that temperatures between 20 to 26°C are preferable for yeast fermentation.
Although sourdoughs are normally prepared without salt, some bakers still opt to use some in their mix. Ganzel’s group found that adding salt to the dough inhibited lactic acid bacteria, including L. sanfranciscensis. This allowed for a faster growth rate of yeast, which were less sensitive to the addition. Thus, salt may be added to encourage fermentation and slow the rate of souring. The bacteria also had a specific range of acidity that encouraged growth. L. sanfranciscensis grew best at pH greater than 4.5, and did not grow well at all under pH 3.8. During the process of sourdough growth and fermentation, the pH drops significantly. Thus, lactic acid production occurs rapidly and stabilizes once pH drops to 3.8 and L. sanfranciscensis is unable to grow. This must be considered in the addition of acetic acid to some commercially-produced sourdoughs-- the pH should match that of a sourdough acidified by L. sanfranciscensis at a pH of 3.8. Furthermore, since S. cerevisiae can tolerate lactic acid but not acetic acid, it is important that the addition of acetic acid to commercially-produced sourdoughs occurs after fermentation so that the bread has a chance to rise. [16]
Conclusion
What does it take to make a perfect sourdough? History would tell us that simply living and baking in San Francisco would do the trick, but we have learned more than that. Lactobacillus sanfranciscensis, a small but mighty microbe, is responsible for the tang that we need to make sourdough bread distinguishable. Though the bacteria is not limited to the San Francisco bay area, it may only be found native to a select 90 countries. Without its symbiotic partner, yeast, the bacteria will simply create a glucose-ridden, sour lump of unrisen bread. But with its mutualistic relationship with the yeast, it can break down flour maltose to contribute glucose to fermentation, creating the fluffy air bubbles we know and love.
L. sanfranciscensis thrives best in a pH range of 3.9 to 6.7, and it is able to grow in up to 4% NaCl. It can tolerate over 160 mmol acetic acid and temperatures between 30°C and 37°C. By contrast, its symbiotic partner C. milleri thrives between 20°C and 27°C. It can handle a larger range of acidity, pH 3.5 to 7, and up to 8% NaCl. However, this yeast’s growth is completely inhibited by 150 mmol acetic acid. Thus, a process that encourages lactic acid production at higher temperatures and fermentation at lower temperatures must be considered in the creation of a proper sourdough. If acetic acid must be added for sourness, it should be done after fermentation to prevent yeast inhibition. Acetic acid may also be produced naturally by the addition of fructose or by increased aeration.
L. sanfranciscensis can be acquired through direct sourcing from another starter or by more organic methods-- from the air or other surfaces. Plant matter (whether vegetable, fruits, or other organic matter) is likely to contain the necessary microbes (L. sanfranciscensis and yeast) for an organic sourdough starter, but may differ in its composition of other microbes. Though L. sanfranciscensis can be combined with other microbes -- or more specifically, other lactic acid bacteria-- to create a sourdough starter, its antimicrobial properties may prevent coexistence with other bacteria. However, a starter rich in microbiota can have a greater volatile compound profile. Thus, a starter rich in L. sanfranciscensis is more convenient and preferable for sourdough creation, but may limit flavor and aroma.
Lactobacillus sanfranciscensis has a quirky history and plays an important role in the production of a well-loved food staple. Small but mighty, this bacterium has evolved to contribute an important role in human lives, and will likely live on in our hearts (and in our sourdough starters) for many generations to come
References
- ↑ : Lee, Sulhee & Jang, Dong-Hun & Jun Choi, Hyuk & Park, Young-Seo. Optimization of Soymilk Fermentation by the Protease-producing Lactobacillus paracasei. Korean Journal of Food Science and Technology. 2013.
- ↑ : Pessione E. Lactic acid bacteria contribution to gut microbiota complexity: lights and shadows. Frontiers in Cellular and Infection Microbiology. 2012
- ↑ : Vogel RF, Pavlovic M, Ehrmann MA, et al. Genomic analysis reveals Lactobacillus sanfranciscensis as stable element in traditional sourdoughs. Microbial Cell Factories. 2011.
- ↑ : L. De Vuyst, S. Van Kerrebroeck, H. Harth, G. Huys, H.-M. Daniel, S. Weckx. Microbial ecology of sourdough fermentations: Diverse or uniform? Food Microbiology, Volume 37, 2014.
- ↑ : Debra Wink. Lactic Acid Fermentation in Sourdough. The Fresh Loaf. 2007
- ↑ : M. Gobbetti, A. Corsetti, Lactobacillus sanfrancisco a key sourdough lactic acid bacterium: a review, Food Microbiology, Volume 14, Issue 2, 1997.
- ↑ : Lhomme, E. ; Orain, S. ; Courcoux, P. ; Onno, B. ; Dousset, X.The predominance of Lactobacillus sanfranciscensis in French organic sourdoughs and its impact on related bread characteristics. International Journal of Food Microbiology, 2015, 213 : 40-48.
- ↑ : P. Damiani, M. Gobbetti, L. Cossignani, A. Corsetti, M.S. Simonetti, J. Rossi. The Sourdough Microflora. Characterization of Hetero- and Homofermentative Lactic Acid Bacteria, Yeasts and Their Interactions on the Basis of the Volatile Compounds Produced. LWT - Food Science and Technology, 1996
- ↑ : Ripari V, Gänzle MG, Berardi E. Evolution of sourdough microbiota in spontaneous sourdoughs started with different plant materials. Int J Food Microbiol. 2016
- ↑ : Biodiversity of Lactobacillus sanfranciscensis strains isolated from five sourdoughs M. Kitahara, S. Sakata and Y. Benno Microbe Division Japan Collection of Microorganisms, RIKEN Bio Resource Center, Wako, Saitama, Japan 2004
- ↑ : Huys G., Daniel HM., De Vuyst L. (2013) Taxonomy and Biodiversity of Sourdough Yeasts and Lactic Acid Bacteria. In: Gobbetti M., Gänzle M. (eds) Handbook on Sourdough Biotechnology. Springer, Boston, MA
- ↑ : Vogel RF, Pavlovic M, Ehrmann MA, et al. Genomic analysis reveals Lactobacillus sanfranciscensis as stable element in traditional sourdoughs. Microbial Cell Factories. 2011
- ↑ : Neubauer H, Glaasker E, Hammes WP, Poolman B, Konings WN. Mechanism of maltose uptake and glucose excretion in Lactobacillus sanfrancisco. Journal of Bacteriology. 1994
- ↑ : Wenge Fu, A.P. Mathews. Lactic acid production from lactose by Lactobacillus plantarum: kinetic model and effects of pH, substrate, and oxygen. Biochemical Engineering Journal, Volume 3, 1999.
- ↑ : Patricia Gadsby and Eric Weeks. The Biology of… Sourdough. Discovery Magazine. 2006
- ↑ : Gänzle MG, Ehmann M, Hammes W. Modeling of Growth of Lactobacillus sanfranciscensis and Candida milleri in Response to Process Parameters of Sourdough Fermentation. Appl Environmental Microbiol. 1998 Jul 1;64(7):2616-23.
Authored for BIOL 238 Microbiology, taught by Joan Slonczewski, 2018, Kenyon College.