Plastic-eating Bacteria: Difference between revisions
| (82 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
<!-- Do not edit this line-->{{Curated}} | <!-- Do not edit this line-->{{Curated}} | ||
By Meheret Ourgessa, student of Professor Joan Slonczweski at Kenyon College | |||
==Introduction== | |||
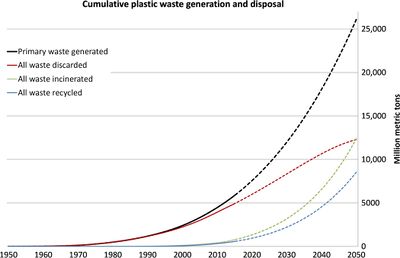
Starting from their introduction in the early 1900s, the use of plastics has grown exponentially, and currently, plastics have become widespread and essential in almost every society on earth to the point that life without plastics is unimaginable for most of us. None of the commonly used plastics are considered biodegradable (meaning that they cannot be dissolved or decomposed by natural agents). As a result, they accumulate, rather than decompose, in landfills or the natural environment.<ref name=plastics/> The continued widespread use of plastics, which does not seem to be decreasing anytime soon, has led to plastic pollution becoming an environmental hazard for which an efficient, easily employable and environmentally-friendly solution is yet to be found.<ref name=pla1>[https://aem.asm.org/content/85/19/e01095-19.full Danso, D., Chow, J., & Streit, W. R. "Plastics: Environmental and Biotechnological Perspectives on Microbial Degradation." 2019b. Applied and Environmental Microbiology, 85(19).]</ref> The main plastic polymers that are produced worldwide are polyurethane, polyethylene, polyamide, polyethylene terephthalate, polystyrene, polyvinyl chloride, and polypropylene.<ref name=pla1/> These and other plastics have become ubiquitous throughout the world, present even in the deepest parts of the world’s oceans such as the Mariana Trench.<ref name=guard1>[https://www.theguardian.com/environment/2018/apr/16/plastic-is-literally-everywhere-the-epidemic-attacking-australias-oceans Readfearn, G. "'Plastic is literally everywhere': the epidemic attacking Australia's oceans" 2018. The Guardian.]</ref><ref name=zme>[https://www.zmescience.com/science/oceanography/plastic-bag-deep-oceans-15052018/ Andrei, M. "Plastics are ubiquitous in the deep ocean -- even in the Mariana Trench." 2018. ZME Science.]</ref> Plastic pollution in oceans continues to increase with plastic debris being found in all major ocean basins, and an estimated 4 to 12 million metric tons of plastic waste generated on land entering the marine environment in 2010 alone.<ref name=plastics/> The first figure (Image One) shows the ever-increasing trend of the amount of plastics used over time with projections into the future.<ref name=plastics/><br> | |||
[[Image:Plastic-Eating Bacteria_PIC1.jpg|thumb|400px|right|Cumulative plastic waste generation and disposal (in million metric tons).</b> Solid lines show historical data from 1950 to 2015; dashed lines show projections of historical trends to 2050 (Image One).<ref name=plastics> [https://advances.sciencemag.org/content/3/7/e1700782.full Geyer, R., Jambeck, J. R., & Law, K. L. "Production, use, and fate of all plastics ever made." 2017. Science Advances, 3(7), e1700782.]</ref>]] | |||
It has been conjectured that the great abundance of plastics has led to the evolution of plastic degrading enzymes such as PETase (PET or polyethylene terephthalate-digesting enzyme) in bacteria.<ref name=PNAS>[https://doi.org/10.1073/pnas.1718804115 Austin, H. P. et. al. "Characterization and engineering of a plastic-degrading aromatic polyesterase." 2018. Proceedings of the National Academy of Sciences, 115(19), E4350–E4357.]</ref> For a long time, much of the plastics that were being produced (basically, all of the plastics mentioned above) were thought to be impossible to degrade via natural agents as the polymers are designed to be durable and stable.<ref name=CEN>[https://cen.acs.org/environment/pollution/Chemistry-solutions-plastic-trash-problem/96/i25 Lemonick, S. "Chemistry may have solutions to our plastic trash problem." 2018. Chemical and Engineering News.]</ref> Furthermore, no enzymes or organisms capable of breaking them down had been identified until 1977, when some lipases (extracellular enzymes that usually cleave esters in oils and fats) were reported to be able to attack ester bonds in some aliphatic polyesters and depolymerize such materials.<ref name=lipase>[https://doi.org/10.1038/270076a0 TOKIWA, Y., & SUZUKI, T. "Hydrolysis of polyesters by lipases." 1917. Nature, 270(5632), 76–78. https://doi.org/10.1038/270076a0]</ref> In recent years, a lot of discoveries later, removal of plastics from the environment using microbes and their enzymes has become a focus of much research.<br> | |||
<br | |||
For the purposes of this page, plastic-eating bacteria are defined as bacteria that can break down and digest plastics such as polyethylene terephthalate (PET), polyethylene (PE) or polyurethane (PU). The recent discovery of <i>Ideonella sakaiensis</i>, which can use the plastic PET as a carbon and energy source,<ref name=ideo>[https://doi.org/10.1126/science.aad6359 Yoshida et al "A bacterium that degrades and assimilates poly(ethylene terephthalate)." 2016. Science, 351(6278), 1196–1199.]</ref> and the following modifications to its genome which have enhanced its degrading capabilities have given new hope for the management of plastic pollution through the utilization of microbes and their enzymes.<ref name=cnn>[https://www.cnn.com/2018/04/16/world/plastic-pollution-science-environment-enzyme-intl/index.html Wilkinson, G. "Scientists hope new enzyme will 'eat' plastic pollution." 2018. CNN.]</ref> A number of other plastic-eating bacteria/microbes have also garnered much attention as the number of enzymes and microorganisms that are capable of breaking down several different types of plastics keeps growing. But, however exciting the applications of these organisms and their enzymes may seem, their capabilities are still limited and require further investigation and modification so that their potential can be fully exploited.<br> | |||
<ref>[ | |||
==<i>Ideonella sakaiensis</i>== | ==<i>Ideonella sakaiensis</i>== | ||
[[Image:Plastic-Eating Bacteria_PIC4.gif|thumb|200px|left|Transmission electron micrograph of an <i>I. sakaiensis</i> cell (Image Two).<ref name=ideoca/>]] | |||
<i> | <i>Ideonella sakaiensis</i> was first discovered in 2016 by Japanese researchers who isolated the novel species from outside a bottle-recycling plant.<ref name=guard2>[https://www.theguardian.com/environment/2016/mar/10/could-a-new-plastic-eating-bacteria-help-combat-this-pollution-scourge Mathiesen, K. "Could a new plastic-eating bacteria help combat this pollution scourge?" 2016. The Guardian.]</ref><ref name=ideo/> The authors identified the species by screening 250 environmental samples at the polyethylene terephthalate (PET) bottle recycling site. <ref name=ideo2>[https://doi.org/10.1126/science.aaf2853 Bornscheuer, U. T. "Feeding on plastic." 2016. Science, 351(6278), 1154–1155.]</ref> At the time, it was the first and only known bacteria species that had naturally evolved to be capable of breaking down and digesting plastic as a carbon and energy source.<ref name=guard2/> Earlier work had shown that the mesophilic fungus <i>Fusarium oxysporum</i> could also produce an enzyme that is able to break down PET into its constituent monomers.<ref name=fungus>[https://doi.org/10.1016/j.bbagen.2015.08.009 Dimarogona, M. et al (2015). "Structural and functional studies of a <i>Fusarium oxysporum</i> cutinase with polyethylene terephthalate modification potential." 2015. Biochimica et Biophysica Acta (BBA) - General Subjects, 1850(11), 2308–2317.]</ref> The discovery of <i>I. sakaiensis</i> was more exciting, however, as bacteria are much easier to harness for industrial uses compared with fungi and other eukaryotes.<ref name=guard3>[https://www.theguardian.com/environment/2018/apr/16/scientists-accidentally-create-mutant-enzyme-that-eats-plastic-bottles Carrington, D. "Scientists accidentally create mutant enzyme that eats plastic bottles." 2018. The Guardian.]</ref> Furthermore, it was shown that the enzyme involved in hydrolyzing PET in <i>I. sakaiensis</i> had as much as 88 times more activity compared to the enzyme found in <i>F. oxysporum</i>.<ref name=ideo/> A hydrolase enzyme isolated from the actinomycete <i>Thermobifida fusca</i> was also shown to effectively depolymerize PET.<ref name=fusca>[https://doi.org/10.1002/marc.200500410 Müller, R. et al "Enzymatic Degradation of Poly(ethylene terephthalate): Rapid Hydrolyse using a Hydrolase from <i>T. fusca</i>." 2005. Macromolecular Rapid Communications, 26(17), 1400–1405.]</ref> But <i>T. Fusca</i> bacteria do not use this enzyme to catabolize PET and the activity of this enzyme is about 120 times lower than that of <i>I. sakaiensis</i>.<ref name=ideo/><br> | ||
<i>I. sakaiensis</i> stain Gram-negative and are aerobic bacteria belonging to the phylum Proteobacteria. The cells are non-spore forming and rod-shaped. The genus <i>Ideonella</i> belongs to the family Comamonadaceae of the class Betaproteobacteria. <i>I. sakaiensis</i> was identified as a representative of a novel species in the genus <i>Ideonella</i> on the basis of its physiological, biochemical, and phylogenetic data, including DNA–DNA relatedness. The DNA–DNA relatedness of <i>I. sakaiensis</i> with its closest phylogenetic neighbors was well below the 70 % cut-off point recommended for the assignment of the strains to the same genomic species. The bacteria grow within the pH range 5.5–9.0 (optimally between pH 7–7.5) and temperature range 15–42 ºC (optimally between 30–37 ºC) and cannot grow with 3% NaCl. The cells of the species are motile and use a polar flagellum to move.<ref name=ideoca>[https://doi.org/10.1099/ijsem.0.001058 Tanasupawat, S., Takehana, T., Yoshida, S., Hiraga, K., & Oda, K. "<i>Ideonella sakaiensis</i> sp. nov., isolated from a microbial consortium that degrades poly(ethylene terephthalate)." 2016. International Journal of Systematic and Evolutionary Microbiology, 66(8), 2813–2818.]</ref> They attach to PET films using appendages which can also grow longer to connect the cells to each other.<ref name=ideo/> Appendages connecting the cell to PET films might also assist in the delivery of secreted enzymes onto the film.<ref name=ideo/> <i>I. sakaiensis</i> can degrade and assimilate PET as a major carbon source for growth. <ref name=ideo/><ref name=ideo2/> The image to the left (Image Two) shows an EM of the bacteria <i>I. sakaiensis</i>. The figure below (Image Three) shows various microscopic pictures and other data that were collected on <i>I. sakaiensis</i> by the original publication on the species.<ref name=ideo/><br> | |||
The researchers who discovered <i>I. sakaiensis</i> identified two enzymes produced by the bacteria that are involved in PET degradation - PETase and MHETase (MHET stands for mono(2-hydroxyethyl) terephthalic acid, the monoester of terephthalic acid and ethylene glycol) - which catabolized PET into its monomers terephthalic acid and ethylene glycol.<ref name=ideo/> These monomers are then used for bacterial metabolism.<ref name=ideo/><ref name=pla1/> Although the PETase enzyme has been described in other bacterial species (mostly belonging to the phylum Actinobacteria), MHETase seems to be unique to the genome of <i>I. sakaiensis</i>.<ref name=pla1/> Originally, the degradation of PET by <i>I. sakaiensis</i> was relatively slow; complete degradation of a small PET film took 6 weeks.<ref name=ideo/><ref name=ideo2/> Recently, researchers were able to genetically modify <i>I. sakaiensis</i> to break down PET faster and also degrade PEF(polyethylene-2,5-furandicarboxylate) which is an emerging bioderived PET replacement with improved properties.<ref name=guard3/><ref name=modeideo>[ https://doi.org/10.1073/pnas.1718804115 Austin, H. P. et al "Characterization and engineering of a plastic-degrading aromatic polyesterase." 2018b. Proceedings of the National Academy of Sciences, 115(19), E4350–E4357.]</ref> The newly modified PETase enzyme is active on the aromatic polyesters (PET and PEF), but not aliphatic polyesters.<ref name=modeideo/> The researchers who engineered this new enzyme concluded that it is likely that significant potential remains for improving its activity through similar protein modifications further.<ref name=modeideo/><br> | [[Image:Plastic-Eating Bacteria_PIC3.jpg|thumb|800px|center|(A) Growth of no. 46 (a consortium of microbes from environmental samples which included <i>I. sakaiensis</i>) on PET film after 20 days. (B) SEM image of degraded PET film after 70 days. The inset shows intact PET film. Scale bar, 0.5 mm. (C) Time course of PET film degradation by no. 46. PET film degradation by I. sakaiensis 201-F6 at 30°C is shown in (D) to (H). The YSV (yeast extract–sodium carbonate–vitamins) medium was changed weekly. (D to F) SEM images of I. sakaiensis cells grown on PET film for 60 hours. Scale bars, 1 μm. Arrowheads in the left panel of (D) indicate contact points of cell appendages and the PET film surface. Magnifications are shown in the right panel. Arrows in (F) indicate appendages between the cell and the PET film surface. (G) SEM image of a degraded PET film surface after washing out adherent cells. The inset shows intact PET film. Scale bar, 1 μm. (H) Time course of PET film degradation by <i>I. sakaiensis</i> (Image Three).<ref name=ideo/>]] | ||
The researchers who discovered <i>I. sakaiensis</i> identified two enzymes produced by the bacteria that are involved in PET degradation - PETase and MHETase (MHET stands for mono(2-hydroxyethyl) terephthalic acid, the monoester of terephthalic acid and ethylene glycol) - which catabolized PET into its monomers terephthalic acid and ethylene glycol.<ref name=ideo/> These monomers are then used for bacterial metabolism.<ref name=ideo/><ref name=pla1/> Although the PETase enzyme has been described in other bacterial species (mostly belonging to the phylum Actinobacteria), MHETase seems to be unique to the genome of <i>I. sakaiensis</i>.<ref name=pla1/> Originally, the degradation of PET by <i>I. sakaiensis</i> was relatively slow; complete degradation of a small PET film took 6 weeks.<ref name=ideo/><ref name=ideo2/> Recently, researchers were able to genetically modify <i>I. sakaiensis</i> to break down PET faster and also degrade PEF(polyethylene-2,5-furandicarboxylate) which is an emerging bioderived PET replacement with improved properties.<ref name=guard3/><ref name=modeideo>[https://doi.org/10.1073/pnas.1718804115 Austin, H. P. et al "Characterization and engineering of a plastic-degrading aromatic polyesterase." 2018b. Proceedings of the National Academy of Sciences, 115(19), E4350–E4357.]</ref> The newly modified PETase enzyme is active on the aromatic polyesters (PET and PEF), but not aliphatic polyesters.<ref name=modeideo/> The researchers who engineered this new enzyme concluded that it is likely that significant potential remains for improving its activity through similar protein modifications further.<ref name=modeideo/><br> | |||
PET was only patented roughly 80 years ago and put into widespread use in the 1970s.<ref name=modeideo/> It is likely that the enzyme system for PET degradation and catabolism in <i>I. sakaiensis</i> appeared only recently, demonstrating the remarkable speed at which microbes adapt and evolve to exploit new substrates.<ref name=modeideo/> In this case, the new substrate was waste from an industrial PET recycling facility. It is thought that a limited number of mutations in a hydrolase, such as PET hydrolytic cutinase, that inherently targets the natural aliphatic polymer cutin (waxy polymer that is one of two main components of the plant cuticle) may have resulted in enhanced selectivity for PET and led to the evolution of the PET degradation pathway in <i>I. sakaiensis</i>.<ref name=ideo/> Genomic analysis has shown that a genomic basis to support the metabolism of MHET analogs was established much earlier than when ancestral PETase proteins were incorporated into the pathway.<ref name=modeideo/> PET enrichment in the habitat of <i>I. sakaiensis</i> is thought to have potentially promoted the selection of a bacterium that might have obtained the necessary set of genes for PET catabolism through lateral gene transfer.<ref name=modeideo/> | PET was only patented roughly 80 years ago and put into widespread use in the 1970s.<ref name=modeideo/> It is likely that the enzyme system for PET degradation and catabolism in <i>I. sakaiensis</i> appeared only recently, demonstrating the remarkable speed at which microbes adapt and evolve to exploit new substrates.<ref name=modeideo/> In this case, the new substrate was waste from an industrial PET recycling facility. It is thought that a limited number of mutations in a hydrolase, such as PET hydrolytic cutinase, that inherently targets the natural aliphatic polymer cutin (waxy polymer that is one of two main components of the plant cuticle) may have resulted in enhanced selectivity for PET and led to the evolution of the PET degradation pathway in <i>I. sakaiensis</i>.<ref name=ideo/> Genomic analysis has shown that a genomic basis to support the metabolism of MHET analogs was established much earlier than when ancestral PETase proteins were incorporated into the pathway.<ref name=modeideo/> PET enrichment in the habitat of <i>I. sakaiensis</i> is thought to have potentially promoted the selection of a bacterium that might have obtained the necessary set of genes for PET catabolism through lateral gene transfer.<ref name=modeideo/> | ||
==Other Plastic-Eating Bacteria== | ==Other Plastic-Eating Bacteria== | ||
Plastics have mainly been introduced since the 1960s. Given the relatively few decades since these polymers became available and widespread, nature has only had a very short time to evolve highly active enzymes. In general, it is thought that the microbial degradation of human-made polymers is a very slow process. The difficulty with the degradation of plastics by microbes mainly stems from the high molecular-weight of the polymers, strong Carbon-Carbon bonds and the extremely hydrophobic surface of plastics, which is very difficult to attack by enzymes.<ref name=pla1/> Nonetheless, a number of bacteria and other microbes have been reported to be able to degrade a variety of plastics(polymers and/or oligomers) or contain enzymes that can.<ref name=pla1/><ref name=micropla2>[https://www.semanticscholar.org/paper/MICROBIAL-DEGRADATION-OF-PLASTIC%3A-A-REVIEW-Kale-Deshmukh/916a631f296fbabcae40daefe4b862a7158cd67b Kale S.K., Deshmukh, A.G., Dudhare, M.S., & Patil, V.B. "MICROBIAL DEGRADATION OF PLASTIC: A REVIEW." 2015. SEMANTICS SCHOLAR]</ref> | Plastics have mainly been introduced since the 1960s. Given the relatively few decades since these polymers became available and widespread, nature has only had a very short time to evolve highly active enzymes. In general, it is thought that the microbial degradation of human-made polymers is a very slow process. The difficulty with the degradation of plastics by microbes mainly stems from the high molecular-weight of the polymers, strong Carbon-Carbon bonds and the extremely hydrophobic surface of plastics, which is very difficult to attack by enzymes.<ref name=pla1/> Nonetheless, a number of bacteria and other microbes have been reported to be able to degrade a variety of plastics(polymers and/or oligomers) or, at least, contain enzymes that can.<ref name=pla1/><ref name=micropla2>[https://www.semanticscholar.org/paper/MICROBIAL-DEGRADATION-OF-PLASTIC%3A-A-REVIEW-Kale-Deshmukh/916a631f296fbabcae40daefe4b862a7158cd67b Kale S.K., Deshmukh, A.G., Dudhare, M.S., & Patil, V.B. "MICROBIAL DEGRADATION OF PLASTIC: A REVIEW." 2015. SEMANTICS SCHOLAR]</ref> The diagram below (Image Four-B) shows a tree containing sequences of enzymes from various organisms known to be able to degrade plastics. | ||
[[Image:Plastic-Eating Bacteria_PIC7.jpg|thumb|400px|left|(A) Electron microscopic images of <i>Comamonas sp.</i>, one of the other bacteria species capable of degrading PET, strain DDHH 01 attached and hydrolyzing PET fibers. Red arrows indicate PET fibers. Black and white arrows indicate bacterial cells. (Top) Transmission electron microscopy image of a PET fiber with attached <i>Comamonas sp.</i> cells. (Middle) Scanning electron microscope image of PET yarn with microcolonies. (Bottom) Closeup of a single cell on the surface of a single PET fiber. (B) Topology of a neighbor-joining tree containing representative sequences of most of the currently known plastic polymer-, oligomer- and monomer-degrading enzymes. The tree is based on amino acid sequence homologies. Overall, 27 known functional and verified enzymes were included in this alignment. This represents the majority of the currently known and biochemically characterized enzymes. PET hydrolases represent the largest fraction of known and studied enzymes (Image Four).<ref name=pla1/>]] | |||
As discussed earlier, researchers have discovered and characterized enzymes from bacteria other than <i>I. sakaiensis</i> that are capable of breaking down PET. Currently, only a few bacteria (and fungi) have been described to be able to partially degrade PET to oligomers or monomers and all known PET hydrolases have relatively low turnover rates. PETases (PET hydrolases - PET hydrolyzing enzymes) represent the best-studied class of enzymes with respect to the hydrolysis of synthetic polymers. Interestingly, the trait for PET degradation appears to be limited to a few bacterial phyla and is mostly found in bacterial isolates that are members of the Gram-positive phylum Actinobacteria. Bacteria that have been found to contain PET hydrolases enzymes include <i>Thermobifida fusca</i>, <i>Bacillus subtilis</i> and species from the genera <i>Thermomonospora</i>. However, none of them have been reported to contain MHETase, the enzyme that breaks down the monomers of PET after it is degraded by a PETase.<ref name=pla1/> Overall, the <i>I. sakaiensis</i> PETase and MHETase enzymes are the best-studied models for PET degradation and it seems that they are the most potent and possibly useful as well.<ref name=pla1/><ref name=ideo/> | As discussed earlier, researchers have discovered and characterized enzymes from bacteria other than <i>I. sakaiensis</i> that are capable of breaking down PET. Currently, only a few bacteria (and fungi) have been described to be able to partially degrade PET to oligomers or monomers and all known PET hydrolases have relatively low turnover rates. PETases (PET hydrolases - PET hydrolyzing enzymes) represent the best-studied class of enzymes with respect to the hydrolysis of synthetic polymers. Interestingly, the trait for PET degradation appears to be limited to a few bacterial phyla and is mostly found in bacterial isolates that are members of the Gram-positive phylum Actinobacteria. Bacteria that have been found to contain PET hydrolases enzymes include <i>Thermobifida fusca</i>, <i>Bacillus subtilis</i> and species from the genera <i>Thermomonospora</i>. However, none of them have been reported to contain MHETase, the enzyme that breaks down the monomers of PET after it is degraded by a PETase.<ref name=pla1/> Overall, the <i>I. sakaiensis</i> PETase and MHETase enzymes are the best-studied models for PET degradation and it seems that they are the most potent and possibly useful as well.<ref name=pla1/><ref name=ideo/> | ||
| Line 50: | Line 35: | ||
Another plastic polymer bacteria have been shown to degrade is polyurethane(PUR/PU).<ref name=pla1/><ref name=front>[https://doi.org/10.3389/fmicb.2020.00404 Espinosa, M. J. C. et al "Toward Biorecycling: Isolation of a Soil Bacterium That Grows on a Polyurethane Oligomer and Monomer." 2020. Frontiers in Microbiology, 11.]</ref> There are a number of different types of polyurethane polymers, but microbes act on only those that have ester links.<ref name=pla1/> Although ether linked polyurethane plastics exist, it seems that no enzymes capable of acting on polyurethane ethers have been described. With respect to bacteria capable of degrading PUR, Gram-negative Betaproteobacteria from the genus <i>Pseudomonas</i> have been most frequently linked with PUR activities.<ref name=pla1/> One Pseudomonas species that has been studied for its PUR-degrading activity is able to grow on a PU-diol solution, a polyurethane oligomer solution, and use it as its sole source of carbon, energy and nitrogen. This bacterial strain was obtained from soil samples and is capable of degrading both, an oligomeric PU and a PU building block(monomer).<ref name=front/> While the list of PU-active bacteria is steadily increasing, various fungal species have also been identified as PU degraders.<ref name=pla1/> | Another plastic polymer bacteria have been shown to degrade is polyurethane(PUR/PU).<ref name=pla1/><ref name=front>[https://doi.org/10.3389/fmicb.2020.00404 Espinosa, M. J. C. et al "Toward Biorecycling: Isolation of a Soil Bacterium That Grows on a Polyurethane Oligomer and Monomer." 2020. Frontiers in Microbiology, 11.]</ref> There are a number of different types of polyurethane polymers, but microbes act on only those that have ester links.<ref name=pla1/> Although ether linked polyurethane plastics exist, it seems that no enzymes capable of acting on polyurethane ethers have been described. With respect to bacteria capable of degrading PUR, Gram-negative Betaproteobacteria from the genus <i>Pseudomonas</i> have been most frequently linked with PUR activities.<ref name=pla1/> One Pseudomonas species that has been studied for its PUR-degrading activity is able to grow on a PU-diol solution, a polyurethane oligomer solution, and use it as its sole source of carbon, energy and nitrogen. This bacterial strain was obtained from soil samples and is capable of degrading both, an oligomeric PU and a PU building block(monomer).<ref name=front/> While the list of PU-active bacteria is steadily increasing, various fungal species have also been identified as PU degraders.<ref name=pla1/> | ||
Polyethylene(PE) has also been associated with microbial degredation. Research performed in polyethylene biodegradation, both with pure strains and complex microbial communities has proved that biodegradation of this material, although slow, is actually happening in nature.<ref name=PE>[https://doi.org/10.1016/j.ibiod.2013.12.014 Restrepo-Flórez, J.-M., Bassi, A., & Thompson, M. R. "Microbial degradation and deterioration of polyethylene – A review." 2014. International Biodeterioration & Biodegradation, 88, 83–90.]</ref> Possible PE degradation has been affiliated with a surprisingly large number of bacterial genera among which are the Gram-negative species in the genera <i>Pseudomonas</i>, <i>Ralstonia</i>, and <i>Stenotrophomonas</i> and also many Gram-positive taxa.<ref name=pla1/> A few studies have also linked PE-degrading microbes with the complex gut microbiomes of invertebrates.<ref name=pla1/> One such study indicated that during short-term exposure, the intestinal microbiome of the larvae of G. mellonella (a greater wax moth) is intricately associated with polyethylene biodegradation in Vivo.<ref name=cater>[Cassone, B. J. et al. "Role of the intestinal microbiome in low-density polyethylene degradation by caterpillar larvae of the greater wax moth, <i>Galleria mellonella</i>." 2020. Proceedings of the Royal Society B: Biological Sciences, 287(1922), 20200112.]</ref> The study identified microorganisms in the genus Acinetobacter that appeared to be involved in this biodegradation process.<ref name=cater/> A number of fungal genera have also been affiliated with assumed PE degradation.<ref name=pla1/> | Polyethylene(PE) has also been associated with microbial degredation. Research performed in polyethylene biodegradation, both with pure strains and complex microbial communities has proved that biodegradation of this material, although slow, is actually happening in nature.<ref name=PE>[https://doi.org/10.1016/j.ibiod.2013.12.014 Restrepo-Flórez, J.-M., Bassi, A., & Thompson, M. R. "Microbial degradation and deterioration of polyethylene – A review." 2014. International Biodeterioration & Biodegradation, 88, 83–90.]</ref> Possible PE degradation has been affiliated with a surprisingly large number of bacterial genera among which are the Gram-negative species in the genera <i>Pseudomonas</i>, <i>Ralstonia</i>, and <i>Stenotrophomonas</i> and also many Gram-positive taxa.<ref name=pla1/> A few studies have also linked PE-degrading microbes with the complex gut microbiomes of invertebrates.<ref name=pla1/> One such study indicated that during short-term exposure, the intestinal microbiome of the larvae of <i>G. mellonella</i> (a greater wax moth) is intricately associated with polyethylene biodegradation in Vivo.<ref name=cater>[https://doi.org/10.1098/rspb.2020.0112 Cassone, B. J. et al. "Role of the intestinal microbiome in low-density polyethylene degradation by caterpillar larvae of the greater wax moth, <i>Galleria mellonella</i>." 2020. Proceedings of the Royal Society B: Biological Sciences, 287(1922), 20200112.]</ref> The study identified microorganisms in the genus Acinetobacter that appeared to be involved in this biodegradation process.<ref name=cater/> A number of fungal genera have also been affiliated with assumed PE degradation.<ref name=pla1/> | ||
Yet Another plastic that has been targeted for bacterial degradation is polyamide. Polyamide(PA) is a polymer of repeating units of aliphatic, semiaromatic, or aromatic molecules linked via amide bonds. Because the monomers for making this polymer can be very versatile, there are many different types of synthetic polyamides, with the most popular being nylon and Kevlar. Although no microorganisms capable of fully degrading the intact high-molecular-weight polymers have been reported, several studies are available on bacteria acting on either linear or cyclic nylon oligomers with short chain lengths. Examples include <i>Arthrobacter sp.</i> and <i>Pseudomonas aeruginosa</i>. The only enzyme that has been reported to act on high-molecular-weight nylon fibers so far originated from a white rot fungus. Some <i>Pseudomonas</i> species have also been reported to utilize 6-aminohexanoate-dimers, oligomers of polyamide, as a sole carbon and nitrogen source.<ref name=pla1/> | |||
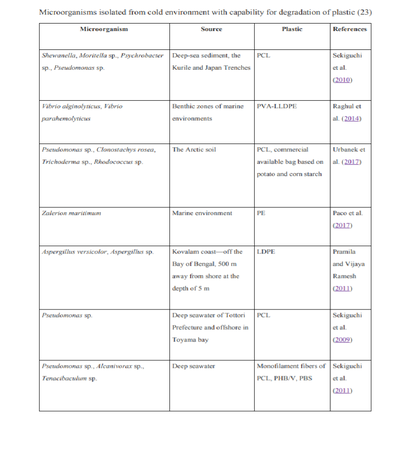
[[Image:Plastic-Eating Bacteria PIC3020.png|thumb|400px|right| Table showing some of the polar microorganisms found to degrade plastics (or having an enzyme that degrades a plastic). The table describes what kind of plastic the microbes are able to degrade and where these microbes were found (Image Five).<ref name=arctic1/>]] | |||
Polystyrene(PS), a synthetic polymer widely used for packaging industries and many daily use articles, has also been a target of possible bacterial degradation. | Polystyrene(PS), a synthetic polymer widely used for packaging industries and many daily use articles, has also been a target of possible bacterial degradation. Unfortunately, much like polyamide, there is no known bacterial enzyme that can degrade the high molecular-weight polystyrene polymer. Styrene itself is able to be used as a carbon source for growth by some microorganisms. Styrene degradation in bacteria is well studied in <i>Pseudomonas</i>, <i>Xanthobacter</i>, <i>Rhodococcus</i>, <i>Corynebacterium</i>, and others. It appears to be a widespread metabolism.<ref name=pla1/> <i>Rhodococcus ruber</i> has been shown to form biofilms on PS and partially degrade it.<ref name=ps>[https://doi.org/10.1080/07388551.2017.1355293 Ho, B. T., Roberts, T. K., & Lucas, S. "An overview on biodegradation of polystyrene and modified polystyrene: the microbial approach." 2017. Critical Reviews in Biotechnology, 38(2), 308–320.]</ref><ref name=pla1/> A biofilter consisting of <i>Brevibacillus sp.</i> has been shown to remove a surprising 3 kg of styrene in a day.<ref name=ps/> In addition, Several bacteria have been reported to form either alone or as members of consortium biofilms on polystyrene films and particles, thereby degrading the polymer.<ref name=pla1/> | ||
In sharp contrast to the huge global production rates of polypropylene and polyvinyl chloride(PVC), hardly any information is available on microbial degradation of either of these important polymers. | In sharp contrast to the huge global production rates of polypropylene and polyvinyl chloride(PVC), hardly any information is available on microbial degradation of either of these important polymers. Though there are a very few reports that describe the degradation of these polymers based on weight loss and using mixed species microbial communities have been published, it is likely that these reports were in part misled by the degradation of the chemical additives rather than the polymer in their cultures. As a result, no defined enzymes or pathways that are responsible for the degradation of either of these two high molecular-weight polymers are yet known.<ref name=pla1/> | ||
Arctic microorganisms may have unique potential for biodegradation of plastics due to the environmental conditions of polar oceans, which differ from other marine ecosystems. | Arctic microorganisms may have unique potential for biodegradation of plastics due to the environmental conditions of polar oceans, which differ from other marine ecosystems. Bacteria from these regions respond quickly to changing environmental patterns. Although there isn’t much research conducted on the interactions between plastic and marine microbiota, the growing amount of plastic waste might force the microorganisms in these environments to adapt to new substrates. Thus, these organisms and their metabolism are prime area for future research on biodegradation of plastics. The table to the right (Image Five) lists some microorganisms isolated from cold environments and able to degrade synthetic polymers(plastics).<ref name=arctic1>[https://doi.org/10.1007/s00253-018-9195-yUrbanek, A. K., Rymowicz, W., & Mirończuk, A. M. "Degradation of plastics and plastic-degrading bacteria in cold marine habitats." 2018. Applied Microbiology and Biotechnology, 102(18), 7669–7678.]</ref> | ||
==Mechanisms of Degradation== | ==Mechanisms of Degradation== | ||
Understanding the mechanisms by which bacteria and other microbes or their enzymes degrade plastic polymers or oligomers can lead to the identification of important enzymes involved in the processes which can be targeted for biotechnological engineering to lead to greater efficiency in microbial degradation of plastics, recycling of plastics and other possible applications. | Understanding the mechanisms by which bacteria and other microbes or their enzymes degrade plastic polymers or oligomers can lead to the identification of important enzymes involved in the processes which can be targeted for biotechnological engineering to lead to greater efficiency in microbial degradation of plastics, recycling of plastics and other possible applications for these microbes and their enzymes. | ||
[[Image:Plastic-Eating Bacteria_PIC5.gif|thumb|300px|left| Representation of a proposed PET degredation mechanism within an <i>I. sakaiensis</i> cell (Image Six).<ref name=ideomech/>]] | |||
One of the most well-studied mechanisms of biodegradation of plastics is the pathway for PET catabolism in <i>I. sakaiensis</i>. It is thought that <i>I. sakaiensis</i> first excretes PETase enzymes (classified as esterase) from the cells onto PET surfaces.<ref name=ideomech/><ref name=ideo/> Appendages that grow extracellularly might be involved in the delivery of PETase onto PET surfaces.<ref name=ideo/> Extracellular PETase then hydrolyzes PET to produce MHET(monoester of ethylene glycol and TPA, both monomers of PET) as the major product and TPA(terephthalic acid) as a minor. The PET hydrolysis products are then transported into the periplasmic space through an outer membrane protein such as porin. MHETase, predicted to be an outer membrane-anchored lipoprotein, hydrolyzes MHET into TPA and EG(ethylene glycol).<ref name=ideomech>[https://doi.org/10.1021/acscatal.8b05171 Taniguchi, I. et al. "Biodegradation of PET: Current Status and Application Aspects." 2019. ACS Catalysis, 9(5), 4089–4105.]</ref> TPA is taken up into the cytoplasm through the TPA transporter coupled with TPA-binding protein and then integrated via protocatechuic acid (PCA) to the tricarboxylic acid (TCA) cycle.<ref name=ideomech/><ref name=ideo/> Ethylene glycol is metabolized via glyoxylic acid to the TCA cycle.<ref name=ideo/> | |||
Interestingly, the expression of the PETase gene was dramatically upregulated in the presence of PET (but not TPA) in a culture of <i>I. sakaiensis</i>, raising the question of which molecule(s) induces its expression. However, TPA does upregulate the expression of a gene cluster that is highly identical with two TPA degradation gene clusters identified in <i>Comamonas sp.</i> strain E6.<ref name=ideo/> The figure (Image Six) to the left illustrates how PET is metabolized in an <i>I. sakaiensis</i> cell. | |||
[[Image:Plastic-Eating Bacteria_PIC6.jpg|thumb|400px|right| Proposed mechanism of degradation of PU oligomers in a <i>Pseudomonas</i> strain (Image Seven).<ref name=front/>]] | |||
Researchers have also proposed a degradation pathway for the oligomers of polyurethane by a <i>Pseudomonas</i> strain. This proposed pathway is summarized in the figure to the right (Image Seven).<ref name=front/> There a number of enzymes involved in this pathway. One of the first enzymes identified to act on PUR was the <i>PueB</i> lipase from <i>Pseudomonas chlororaphis</i>, a different Pseudomonas species.<ref name=pla1/> This organism codes for at least one additional enzyme active on PU, which was designated <i>PueA</i>. Both enzymes are lipases (which usually catalyze the ester-linked lipids in other cells) and similar to PET degradation with microbes, PU is degraded by the secreted hydrolases, and the degradation is tightly regulated. As mentioned previously, enzymes and microbes described as capable of degrading organisms were all acting on ester-linked PU. There have yet to be enzymes described to act on polyurethane ethers in the scientific literature.<ref name=pla1/> | |||
To date, no microorganisms capable of degrading the intact high-molecular-weight polyamide polymer have been reported, but there are many bacteria that act on the oligomers of polyamide. Three main enzymes have been reported as essential for the initial hydrolysis of cyclic and linear 6-aminohexanoate oligomers. The first one is a cyclic-dimer hydrolase (<i>NylA</i>), the second a dimer hydrolase (<i>NylB</i>), and the third an endo-type oligomer hydrolase (<i>NylC</i>). Once the oligomers are hydrolyzed, the monomers are metabolized by different aminotransferases.<ref name=pla1/> | |||
As with polyamides, there are no known microorganisms capable of degrading the high-molecular-weight polystyrene polymer and neither are there any enzymes, but there are many microbes that degrade the oligomers of the plastic.<ref name=pla1/> Under aerobic conditions, styrene is oxidized by two different pathways. The first one involves attacking the vinyl side chain and the second one is through attacking a rather nonspecific aromatic ring, thereby forming primarily the intermediates 3-vinylcatechol, phenylacetic acid, and 2-phenylethanol. These intermediates are channeled into the Krebs cycle after ring cleavage. The degradation of the vinyl side chain involves the action of three key enzymes, a styrene monooxygenase complex(<i>styA</i> and <i>styB</i>), a styrene oxide isomerase(<i>styC</i>), and a phenylacetaldehyde dehydrogenase(<i>styD</i>). The respective genes for side-chain oxygenation are frequently located in a single conserved gene cluster, often designated <i>styABC(D)</i>. The styrene monooxygenase attacks the vinyl side chain to release epoxystyrene, which is then subjected to isomerization to form phenylacetaldehyde. Phenylacetaldehyde is then oxidized to phenylacetic acid through the involvement of a dehydrogenase.<ref name=pla1/> In <i>P. putida</i>, the phenylacetic acid is activated to phenylacetyl-coenzyme A (CoA) and then subjected to β-oxidation to yield acetyl-CoA, which is directly fed into the Krebs cycle.<ref name=putida>[https://doi.org/10.1186/1471-2180-11-229 O' Leary, N.D. et al. "Regulation of phenylacetic acid uptake is σ54 dependent in Pseudomonas putidaCA-3." 2011. BMC Microbiol 11, 229.]</ref><ref name=pla1/> The expression of the conserved cluster is regulated through either a two-component regulatory system or LysR-type regulators.<ref name=pla1/> | |||
Research performed on PE degradation by microbes so far is mainly of descriptive nature, with a few studies devoted to polyethylene degradation mechanisms or the isolation of enzymes belonging related to this process. However, these findings are limited and further evidence is required to identify the complete mechanisms for polyethylene degradation.<ref name=PE/> Similarly, the few studies that describe the degradation of polyvinyl chloride and polypropylene by microbial communities do not detail the enzymes and pathways involved in the process.<ref name=pla1/> | |||
==Significance, Possible Applications and Limitations== | |||
[[Image: Plastic-Eating Bacteria_PIC8.jpg|thumb|600px|center|Global production, use, and fate of polymer resins, synthetic fibers, and additives (1950 to 2015; in million metric tons).<ref name=plastics/> This flow chart demonstrates the urgent need for an effective and environmentally friendly solution for plastic pollution (Image Eight).]] | |||
As the flowchart above (Image Eight) shows, plastic pollution is a major problem that needs an effective solution soon. The majority of plastics that are produced are discarded into the environment while a smaller percentage is continually used and even less is recycled.<ref name=plastics/> The hope is that genetically-engineered plastic-degrading bacteria may help recycle and/or degrade (in an environmentally friendly way) the produced plastics. There are many different ways plastic-degrading bacteria can be applied to improve plastic recycling or decomposition and decrease or curb pollution. Current possible applications of microbial degradation of plastics are limited, but these microbes may eventually be used to completely degrade certain plastic polymers or their oligomers and monomers.<ref name=arctic1/> For example, The assimilation of PET by <i>I. sakaiensis</i> bacteria may be advantageous for removing this plastic material from the environment.<ref name=ideo2/> Plastic-degrading microbes could also be applied in the recycling process. In PET degradation, if the terephthalic acid could be isolated and reused, this could provide huge savings in the production of new polymer without the need for petrol-based starting materials.<ref name=ideo2/> | |||
Another potential route of application is the isolation of the genes encoding enzymes for biodegradation which can then be integrated into common production strains via metabolic engineering or the use of enzyme cascade systems.<ref name=ideo2/><ref name=front/> Alternatively, genetic sequences of these enzymes could be used to mass-produce the enzymes themselves and use them to directly degrade plastic polymers.<ref name=ideomech/><ref name=arctic1/> Furthermore, engineering microorganisms that would produce high-value compounds from plastic waste is a future challenge and would contribute to an improved circular use of plastics. Monomers and oligomers formed after the degradation could be used to build value-added products or even new polymers that are biodegradable.<ref name=pla1/> In addition, the development of highly active enzymes to be used textile industries and in detergents(cellulase is already currently used as one of the additives to detergents for cotton fibers) could already significantly reduce annual plastic pollution.<ref name=pla1/><ref name=arctic1/> | |||
The uniquely cold-adapted enzymes of polar microorganisms provide numerous opportunities for biotechnological exploitation and give new insights into a wide range of applied issues such as plastic pollution. Currently, enzymes from psychrophilic microorganisms are raising interest for many industrial applications due to ongoing attempts to decrease energy demand. The lower temperature needed for growth at which enzymatic activity is maintained may be a huge advantage in the degradation process due to reduction of electric energy usage for heating.<ref name=arctic1/> | |||
== | There are many limiting factors in the application of plastic-degrading bacteria. Firstly, microbial plastic degradation of intact high-molecular-weight polymers seems to be limited to only polyethylene terephthalate.<ref name=pla1/> The microbial mechanisms that degrade other plastics are only limited to breaking down oligomers and monomers. And even with PET degradation the crystallinity and durability of the intact polymer limits the process.<ref name=pla1/> This is also often true for many other bacterial mechanisms for breaking down plastics.<ref name=front/> Another limiting factor that needs to be improved upon is bacterial survivability in plastic polluted environments. With <i>I. sakaiensis</i>, headway has already been made as genetic engineering has enabled the bacterial to survive in usual areas where plastic waste is located so that the bacteria can degrade the plastic waste optimally.<ref name=pla1/> The turn over rate of enzymes involved in plastic degradation and the overall rate of plastic degradation by microorganisms also needs to be improved before microbes and their enzymes can be applied to curb plastic pollution. Currently, the majority of bacteria that degrade plastics take as much as several weeks to fully or substantially break down small amounts of plastic films.<ref name=fusca/><ref name=ideo/> Again, important headway is being made on this front with the bacterium <i>I. sakaiensis</i> of which the PETase enzyme was recently genetically modified to increase degradation activity by 14-fold at 40 °C.<ref>[https://doi.org/10.2139/ssrn.3194556 Widyastuti, G. "Genetic Engineered Ideonella Sakaiensis Bacteria: A Solution of the Legendary Plastic Waste Problem." 2018. SSRN Electronic Journal.]</ref> | ||
It is important to note that since commercially available polymers and films are often used as substrates, they contain additives, plasticizers, and other biodegradable impurities (for example, phthalates), which are much more easily broken down than the actual backbone. This, therefore, interferes with the results studies show for plastic degradation by microbes or their enzymes and frequently leads to the identification of false positives. Also, often the enzyme activities that are reported are based on clearing zones in agar plates. However, these assays are not fully reliable. For these reasons, the overall methodology linked to the analysis of microbial plastic degradation needs to be standardized and optimized.<ref name=pla1/> | |||
==Conclusion== | ==Conclusion== | ||
All this information about plastic-degrading bacteria points to the untapped potential of bacteria and their enzymes in curbing and decreasing plastic pollution in different natural environments. More research needs to be done to identify the enzymes, mechanisms, and especially microbial interactions involved in biodegradation. This can go a long way to improve the microbial degradation of plastics and lead to refined processes for applications. Currently, possible prime research areas include investigation of the degradation of plastics by polar microbes, elucidating microbial interactions involved in the degradation of different plastic polymers, and developing better methods to properly measure the amount of plastics broken down. Another area of future research that looks promising is the genetic modification of I. sakaiensis’ PETase enzyme to yield a more efficient hydrolase. At this time, the most effective way to break down plastics involves exposure to UV light together with mechanical disruption caused by waves and winds or grinding on marine rocks and sediments, which eventually breaks larger plastics into smaller pieces of micro- and nanoplastics.<ref name=pla1/> This method has its own limitations (including not being environmentally friendly<ref name=pla1/>) and microbial degradation of plastics may well be the method of choice in our future endeavors to recycle and/or break down the substantial amount of plastics already polluting almost all of the world’s natural environment. | |||
==References== | ==References== | ||
<references /> | <references /> | ||
<br><br>Authored for BIOL 238 Microbiology, taught by [mailto:slonczewski@kenyon.edu Joan Slonczewski], | <br><br>Authored for BIOL 238 Microbiology, taught by [mailto:slonczewski@kenyon.edu Joan Slonczewski], 2020, [http://www.kenyon.edu/index.xml Kenyon College]. | ||
Latest revision as of 19:26, 15 May 2020
By Meheret Ourgessa, student of Professor Joan Slonczweski at Kenyon College
Introduction
Starting from their introduction in the early 1900s, the use of plastics has grown exponentially, and currently, plastics have become widespread and essential in almost every society on earth to the point that life without plastics is unimaginable for most of us. None of the commonly used plastics are considered biodegradable (meaning that they cannot be dissolved or decomposed by natural agents). As a result, they accumulate, rather than decompose, in landfills or the natural environment.[1] The continued widespread use of plastics, which does not seem to be decreasing anytime soon, has led to plastic pollution becoming an environmental hazard for which an efficient, easily employable and environmentally-friendly solution is yet to be found.[2] The main plastic polymers that are produced worldwide are polyurethane, polyethylene, polyamide, polyethylene terephthalate, polystyrene, polyvinyl chloride, and polypropylene.[2] These and other plastics have become ubiquitous throughout the world, present even in the deepest parts of the world’s oceans such as the Mariana Trench.[3][4] Plastic pollution in oceans continues to increase with plastic debris being found in all major ocean basins, and an estimated 4 to 12 million metric tons of plastic waste generated on land entering the marine environment in 2010 alone.[1] The first figure (Image One) shows the ever-increasing trend of the amount of plastics used over time with projections into the future.[1]
It has been conjectured that the great abundance of plastics has led to the evolution of plastic degrading enzymes such as PETase (PET or polyethylene terephthalate-digesting enzyme) in bacteria.[5] For a long time, much of the plastics that were being produced (basically, all of the plastics mentioned above) were thought to be impossible to degrade via natural agents as the polymers are designed to be durable and stable.[6] Furthermore, no enzymes or organisms capable of breaking them down had been identified until 1977, when some lipases (extracellular enzymes that usually cleave esters in oils and fats) were reported to be able to attack ester bonds in some aliphatic polyesters and depolymerize such materials.[7] In recent years, a lot of discoveries later, removal of plastics from the environment using microbes and their enzymes has become a focus of much research.
For the purposes of this page, plastic-eating bacteria are defined as bacteria that can break down and digest plastics such as polyethylene terephthalate (PET), polyethylene (PE) or polyurethane (PU). The recent discovery of Ideonella sakaiensis, which can use the plastic PET as a carbon and energy source,[8] and the following modifications to its genome which have enhanced its degrading capabilities have given new hope for the management of plastic pollution through the utilization of microbes and their enzymes.[9] A number of other plastic-eating bacteria/microbes have also garnered much attention as the number of enzymes and microorganisms that are capable of breaking down several different types of plastics keeps growing. But, however exciting the applications of these organisms and their enzymes may seem, their capabilities are still limited and require further investigation and modification so that their potential can be fully exploited.
Ideonella sakaiensis
Ideonella sakaiensis was first discovered in 2016 by Japanese researchers who isolated the novel species from outside a bottle-recycling plant.[11][8] The authors identified the species by screening 250 environmental samples at the polyethylene terephthalate (PET) bottle recycling site. [12] At the time, it was the first and only known bacteria species that had naturally evolved to be capable of breaking down and digesting plastic as a carbon and energy source.[11] Earlier work had shown that the mesophilic fungus Fusarium oxysporum could also produce an enzyme that is able to break down PET into its constituent monomers.[13] The discovery of I. sakaiensis was more exciting, however, as bacteria are much easier to harness for industrial uses compared with fungi and other eukaryotes.[14] Furthermore, it was shown that the enzyme involved in hydrolyzing PET in I. sakaiensis had as much as 88 times more activity compared to the enzyme found in F. oxysporum.[8] A hydrolase enzyme isolated from the actinomycete Thermobifida fusca was also shown to effectively depolymerize PET.[15] But T. Fusca bacteria do not use this enzyme to catabolize PET and the activity of this enzyme is about 120 times lower than that of I. sakaiensis.[8]
I. sakaiensis stain Gram-negative and are aerobic bacteria belonging to the phylum Proteobacteria. The cells are non-spore forming and rod-shaped. The genus Ideonella belongs to the family Comamonadaceae of the class Betaproteobacteria. I. sakaiensis was identified as a representative of a novel species in the genus Ideonella on the basis of its physiological, biochemical, and phylogenetic data, including DNA–DNA relatedness. The DNA–DNA relatedness of I. sakaiensis with its closest phylogenetic neighbors was well below the 70 % cut-off point recommended for the assignment of the strains to the same genomic species. The bacteria grow within the pH range 5.5–9.0 (optimally between pH 7–7.5) and temperature range 15–42 ºC (optimally between 30–37 ºC) and cannot grow with 3% NaCl. The cells of the species are motile and use a polar flagellum to move.[10] They attach to PET films using appendages which can also grow longer to connect the cells to each other.[8] Appendages connecting the cell to PET films might also assist in the delivery of secreted enzymes onto the film.[8] I. sakaiensis can degrade and assimilate PET as a major carbon source for growth. [8][12] The image to the left (Image Two) shows an EM of the bacteria I. sakaiensis. The figure below (Image Three) shows various microscopic pictures and other data that were collected on I. sakaiensis by the original publication on the species.[8]

The researchers who discovered I. sakaiensis identified two enzymes produced by the bacteria that are involved in PET degradation - PETase and MHETase (MHET stands for mono(2-hydroxyethyl) terephthalic acid, the monoester of terephthalic acid and ethylene glycol) - which catabolized PET into its monomers terephthalic acid and ethylene glycol.[8] These monomers are then used for bacterial metabolism.[8][2] Although the PETase enzyme has been described in other bacterial species (mostly belonging to the phylum Actinobacteria), MHETase seems to be unique to the genome of I. sakaiensis.[2] Originally, the degradation of PET by I. sakaiensis was relatively slow; complete degradation of a small PET film took 6 weeks.[8][12] Recently, researchers were able to genetically modify I. sakaiensis to break down PET faster and also degrade PEF(polyethylene-2,5-furandicarboxylate) which is an emerging bioderived PET replacement with improved properties.[14][16] The newly modified PETase enzyme is active on the aromatic polyesters (PET and PEF), but not aliphatic polyesters.[16] The researchers who engineered this new enzyme concluded that it is likely that significant potential remains for improving its activity through similar protein modifications further.[16]
PET was only patented roughly 80 years ago and put into widespread use in the 1970s.[16] It is likely that the enzyme system for PET degradation and catabolism in I. sakaiensis appeared only recently, demonstrating the remarkable speed at which microbes adapt and evolve to exploit new substrates.[16] In this case, the new substrate was waste from an industrial PET recycling facility. It is thought that a limited number of mutations in a hydrolase, such as PET hydrolytic cutinase, that inherently targets the natural aliphatic polymer cutin (waxy polymer that is one of two main components of the plant cuticle) may have resulted in enhanced selectivity for PET and led to the evolution of the PET degradation pathway in I. sakaiensis.[8] Genomic analysis has shown that a genomic basis to support the metabolism of MHET analogs was established much earlier than when ancestral PETase proteins were incorporated into the pathway.[16] PET enrichment in the habitat of I. sakaiensis is thought to have potentially promoted the selection of a bacterium that might have obtained the necessary set of genes for PET catabolism through lateral gene transfer.[16]
Other Plastic-Eating Bacteria
Plastics have mainly been introduced since the 1960s. Given the relatively few decades since these polymers became available and widespread, nature has only had a very short time to evolve highly active enzymes. In general, it is thought that the microbial degradation of human-made polymers is a very slow process. The difficulty with the degradation of plastics by microbes mainly stems from the high molecular-weight of the polymers, strong Carbon-Carbon bonds and the extremely hydrophobic surface of plastics, which is very difficult to attack by enzymes.[2] Nonetheless, a number of bacteria and other microbes have been reported to be able to degrade a variety of plastics(polymers and/or oligomers) or, at least, contain enzymes that can.[2][17] The diagram below (Image Four-B) shows a tree containing sequences of enzymes from various organisms known to be able to degrade plastics.
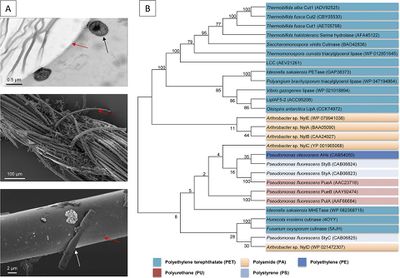
As discussed earlier, researchers have discovered and characterized enzymes from bacteria other than I. sakaiensis that are capable of breaking down PET. Currently, only a few bacteria (and fungi) have been described to be able to partially degrade PET to oligomers or monomers and all known PET hydrolases have relatively low turnover rates. PETases (PET hydrolases - PET hydrolyzing enzymes) represent the best-studied class of enzymes with respect to the hydrolysis of synthetic polymers. Interestingly, the trait for PET degradation appears to be limited to a few bacterial phyla and is mostly found in bacterial isolates that are members of the Gram-positive phylum Actinobacteria. Bacteria that have been found to contain PET hydrolases enzymes include Thermobifida fusca, Bacillus subtilis and species from the genera Thermomonospora. However, none of them have been reported to contain MHETase, the enzyme that breaks down the monomers of PET after it is degraded by a PETase.[2] Overall, the I. sakaiensis PETase and MHETase enzymes are the best-studied models for PET degradation and it seems that they are the most potent and possibly useful as well.[2][8]
Another plastic polymer bacteria have been shown to degrade is polyurethane(PUR/PU).[2][18] There are a number of different types of polyurethane polymers, but microbes act on only those that have ester links.[2] Although ether linked polyurethane plastics exist, it seems that no enzymes capable of acting on polyurethane ethers have been described. With respect to bacteria capable of degrading PUR, Gram-negative Betaproteobacteria from the genus Pseudomonas have been most frequently linked with PUR activities.[2] One Pseudomonas species that has been studied for its PUR-degrading activity is able to grow on a PU-diol solution, a polyurethane oligomer solution, and use it as its sole source of carbon, energy and nitrogen. This bacterial strain was obtained from soil samples and is capable of degrading both, an oligomeric PU and a PU building block(monomer).[18] While the list of PU-active bacteria is steadily increasing, various fungal species have also been identified as PU degraders.[2]
Polyethylene(PE) has also been associated with microbial degredation. Research performed in polyethylene biodegradation, both with pure strains and complex microbial communities has proved that biodegradation of this material, although slow, is actually happening in nature.[19] Possible PE degradation has been affiliated with a surprisingly large number of bacterial genera among which are the Gram-negative species in the genera Pseudomonas, Ralstonia, and Stenotrophomonas and also many Gram-positive taxa.[2] A few studies have also linked PE-degrading microbes with the complex gut microbiomes of invertebrates.[2] One such study indicated that during short-term exposure, the intestinal microbiome of the larvae of G. mellonella (a greater wax moth) is intricately associated with polyethylene biodegradation in Vivo.[20] The study identified microorganisms in the genus Acinetobacter that appeared to be involved in this biodegradation process.[20] A number of fungal genera have also been affiliated with assumed PE degradation.[2]
Yet Another plastic that has been targeted for bacterial degradation is polyamide. Polyamide(PA) is a polymer of repeating units of aliphatic, semiaromatic, or aromatic molecules linked via amide bonds. Because the monomers for making this polymer can be very versatile, there are many different types of synthetic polyamides, with the most popular being nylon and Kevlar. Although no microorganisms capable of fully degrading the intact high-molecular-weight polymers have been reported, several studies are available on bacteria acting on either linear or cyclic nylon oligomers with short chain lengths. Examples include Arthrobacter sp. and Pseudomonas aeruginosa. The only enzyme that has been reported to act on high-molecular-weight nylon fibers so far originated from a white rot fungus. Some Pseudomonas species have also been reported to utilize 6-aminohexanoate-dimers, oligomers of polyamide, as a sole carbon and nitrogen source.[2]
Polystyrene(PS), a synthetic polymer widely used for packaging industries and many daily use articles, has also been a target of possible bacterial degradation. Unfortunately, much like polyamide, there is no known bacterial enzyme that can degrade the high molecular-weight polystyrene polymer. Styrene itself is able to be used as a carbon source for growth by some microorganisms. Styrene degradation in bacteria is well studied in Pseudomonas, Xanthobacter, Rhodococcus, Corynebacterium, and others. It appears to be a widespread metabolism.[2] Rhodococcus ruber has been shown to form biofilms on PS and partially degrade it.[22][2] A biofilter consisting of Brevibacillus sp. has been shown to remove a surprising 3 kg of styrene in a day.[22] In addition, Several bacteria have been reported to form either alone or as members of consortium biofilms on polystyrene films and particles, thereby degrading the polymer.[2]
In sharp contrast to the huge global production rates of polypropylene and polyvinyl chloride(PVC), hardly any information is available on microbial degradation of either of these important polymers. Though there are a very few reports that describe the degradation of these polymers based on weight loss and using mixed species microbial communities have been published, it is likely that these reports were in part misled by the degradation of the chemical additives rather than the polymer in their cultures. As a result, no defined enzymes or pathways that are responsible for the degradation of either of these two high molecular-weight polymers are yet known.[2]
Arctic microorganisms may have unique potential for biodegradation of plastics due to the environmental conditions of polar oceans, which differ from other marine ecosystems. Bacteria from these regions respond quickly to changing environmental patterns. Although there isn’t much research conducted on the interactions between plastic and marine microbiota, the growing amount of plastic waste might force the microorganisms in these environments to adapt to new substrates. Thus, these organisms and their metabolism are prime area for future research on biodegradation of plastics. The table to the right (Image Five) lists some microorganisms isolated from cold environments and able to degrade synthetic polymers(plastics).[21]
Mechanisms of Degradation
Understanding the mechanisms by which bacteria and other microbes or their enzymes degrade plastic polymers or oligomers can lead to the identification of important enzymes involved in the processes which can be targeted for biotechnological engineering to lead to greater efficiency in microbial degradation of plastics, recycling of plastics and other possible applications for these microbes and their enzymes.
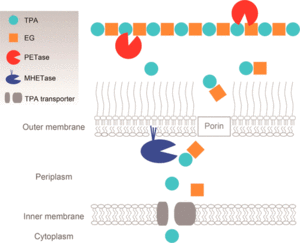
One of the most well-studied mechanisms of biodegradation of plastics is the pathway for PET catabolism in I. sakaiensis. It is thought that I. sakaiensis first excretes PETase enzymes (classified as esterase) from the cells onto PET surfaces.[23][8] Appendages that grow extracellularly might be involved in the delivery of PETase onto PET surfaces.[8] Extracellular PETase then hydrolyzes PET to produce MHET(monoester of ethylene glycol and TPA, both monomers of PET) as the major product and TPA(terephthalic acid) as a minor. The PET hydrolysis products are then transported into the periplasmic space through an outer membrane protein such as porin. MHETase, predicted to be an outer membrane-anchored lipoprotein, hydrolyzes MHET into TPA and EG(ethylene glycol).[23] TPA is taken up into the cytoplasm through the TPA transporter coupled with TPA-binding protein and then integrated via protocatechuic acid (PCA) to the tricarboxylic acid (TCA) cycle.[23][8] Ethylene glycol is metabolized via glyoxylic acid to the TCA cycle.[8]
Interestingly, the expression of the PETase gene was dramatically upregulated in the presence of PET (but not TPA) in a culture of I. sakaiensis, raising the question of which molecule(s) induces its expression. However, TPA does upregulate the expression of a gene cluster that is highly identical with two TPA degradation gene clusters identified in Comamonas sp. strain E6.[8] The figure (Image Six) to the left illustrates how PET is metabolized in an I. sakaiensis cell.

Researchers have also proposed a degradation pathway for the oligomers of polyurethane by a Pseudomonas strain. This proposed pathway is summarized in the figure to the right (Image Seven).[18] There a number of enzymes involved in this pathway. One of the first enzymes identified to act on PUR was the PueB lipase from Pseudomonas chlororaphis, a different Pseudomonas species.[2] This organism codes for at least one additional enzyme active on PU, which was designated PueA. Both enzymes are lipases (which usually catalyze the ester-linked lipids in other cells) and similar to PET degradation with microbes, PU is degraded by the secreted hydrolases, and the degradation is tightly regulated. As mentioned previously, enzymes and microbes described as capable of degrading organisms were all acting on ester-linked PU. There have yet to be enzymes described to act on polyurethane ethers in the scientific literature.[2]
To date, no microorganisms capable of degrading the intact high-molecular-weight polyamide polymer have been reported, but there are many bacteria that act on the oligomers of polyamide. Three main enzymes have been reported as essential for the initial hydrolysis of cyclic and linear 6-aminohexanoate oligomers. The first one is a cyclic-dimer hydrolase (NylA), the second a dimer hydrolase (NylB), and the third an endo-type oligomer hydrolase (NylC). Once the oligomers are hydrolyzed, the monomers are metabolized by different aminotransferases.[2]
As with polyamides, there are no known microorganisms capable of degrading the high-molecular-weight polystyrene polymer and neither are there any enzymes, but there are many microbes that degrade the oligomers of the plastic.[2] Under aerobic conditions, styrene is oxidized by two different pathways. The first one involves attacking the vinyl side chain and the second one is through attacking a rather nonspecific aromatic ring, thereby forming primarily the intermediates 3-vinylcatechol, phenylacetic acid, and 2-phenylethanol. These intermediates are channeled into the Krebs cycle after ring cleavage. The degradation of the vinyl side chain involves the action of three key enzymes, a styrene monooxygenase complex(styA and styB), a styrene oxide isomerase(styC), and a phenylacetaldehyde dehydrogenase(styD). The respective genes for side-chain oxygenation are frequently located in a single conserved gene cluster, often designated styABC(D). The styrene monooxygenase attacks the vinyl side chain to release epoxystyrene, which is then subjected to isomerization to form phenylacetaldehyde. Phenylacetaldehyde is then oxidized to phenylacetic acid through the involvement of a dehydrogenase.[2] In P. putida, the phenylacetic acid is activated to phenylacetyl-coenzyme A (CoA) and then subjected to β-oxidation to yield acetyl-CoA, which is directly fed into the Krebs cycle.[24][2] The expression of the conserved cluster is regulated through either a two-component regulatory system or LysR-type regulators.[2]
Research performed on PE degradation by microbes so far is mainly of descriptive nature, with a few studies devoted to polyethylene degradation mechanisms or the isolation of enzymes belonging related to this process. However, these findings are limited and further evidence is required to identify the complete mechanisms for polyethylene degradation.[19] Similarly, the few studies that describe the degradation of polyvinyl chloride and polypropylene by microbial communities do not detail the enzymes and pathways involved in the process.[2]
Significance, Possible Applications and Limitations
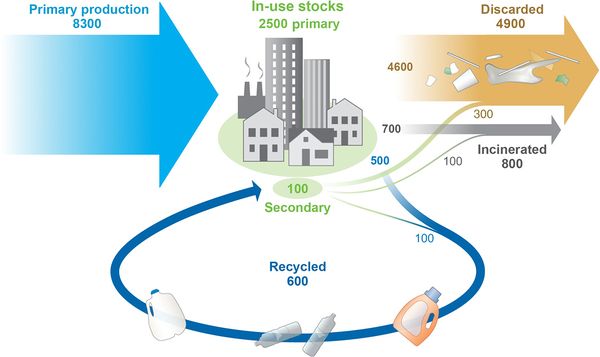
As the flowchart above (Image Eight) shows, plastic pollution is a major problem that needs an effective solution soon. The majority of plastics that are produced are discarded into the environment while a smaller percentage is continually used and even less is recycled.[1] The hope is that genetically-engineered plastic-degrading bacteria may help recycle and/or degrade (in an environmentally friendly way) the produced plastics. There are many different ways plastic-degrading bacteria can be applied to improve plastic recycling or decomposition and decrease or curb pollution. Current possible applications of microbial degradation of plastics are limited, but these microbes may eventually be used to completely degrade certain plastic polymers or their oligomers and monomers.[21] For example, The assimilation of PET by I. sakaiensis bacteria may be advantageous for removing this plastic material from the environment.[12] Plastic-degrading microbes could also be applied in the recycling process. In PET degradation, if the terephthalic acid could be isolated and reused, this could provide huge savings in the production of new polymer without the need for petrol-based starting materials.[12]
Another potential route of application is the isolation of the genes encoding enzymes for biodegradation which can then be integrated into common production strains via metabolic engineering or the use of enzyme cascade systems.[12][18] Alternatively, genetic sequences of these enzymes could be used to mass-produce the enzymes themselves and use them to directly degrade plastic polymers.[23][21] Furthermore, engineering microorganisms that would produce high-value compounds from plastic waste is a future challenge and would contribute to an improved circular use of plastics. Monomers and oligomers formed after the degradation could be used to build value-added products or even new polymers that are biodegradable.[2] In addition, the development of highly active enzymes to be used textile industries and in detergents(cellulase is already currently used as one of the additives to detergents for cotton fibers) could already significantly reduce annual plastic pollution.[2][21]
The uniquely cold-adapted enzymes of polar microorganisms provide numerous opportunities for biotechnological exploitation and give new insights into a wide range of applied issues such as plastic pollution. Currently, enzymes from psychrophilic microorganisms are raising interest for many industrial applications due to ongoing attempts to decrease energy demand. The lower temperature needed for growth at which enzymatic activity is maintained may be a huge advantage in the degradation process due to reduction of electric energy usage for heating.[21]
There are many limiting factors in the application of plastic-degrading bacteria. Firstly, microbial plastic degradation of intact high-molecular-weight polymers seems to be limited to only polyethylene terephthalate.[2] The microbial mechanisms that degrade other plastics are only limited to breaking down oligomers and monomers. And even with PET degradation the crystallinity and durability of the intact polymer limits the process.[2] This is also often true for many other bacterial mechanisms for breaking down plastics.[18] Another limiting factor that needs to be improved upon is bacterial survivability in plastic polluted environments. With I. sakaiensis, headway has already been made as genetic engineering has enabled the bacterial to survive in usual areas where plastic waste is located so that the bacteria can degrade the plastic waste optimally.[2] The turn over rate of enzymes involved in plastic degradation and the overall rate of plastic degradation by microorganisms also needs to be improved before microbes and their enzymes can be applied to curb plastic pollution. Currently, the majority of bacteria that degrade plastics take as much as several weeks to fully or substantially break down small amounts of plastic films.[15][8] Again, important headway is being made on this front with the bacterium I. sakaiensis of which the PETase enzyme was recently genetically modified to increase degradation activity by 14-fold at 40 °C.[25]
It is important to note that since commercially available polymers and films are often used as substrates, they contain additives, plasticizers, and other biodegradable impurities (for example, phthalates), which are much more easily broken down than the actual backbone. This, therefore, interferes with the results studies show for plastic degradation by microbes or their enzymes and frequently leads to the identification of false positives. Also, often the enzyme activities that are reported are based on clearing zones in agar plates. However, these assays are not fully reliable. For these reasons, the overall methodology linked to the analysis of microbial plastic degradation needs to be standardized and optimized.[2]
Conclusion
All this information about plastic-degrading bacteria points to the untapped potential of bacteria and their enzymes in curbing and decreasing plastic pollution in different natural environments. More research needs to be done to identify the enzymes, mechanisms, and especially microbial interactions involved in biodegradation. This can go a long way to improve the microbial degradation of plastics and lead to refined processes for applications. Currently, possible prime research areas include investigation of the degradation of plastics by polar microbes, elucidating microbial interactions involved in the degradation of different plastic polymers, and developing better methods to properly measure the amount of plastics broken down. Another area of future research that looks promising is the genetic modification of I. sakaiensis’ PETase enzyme to yield a more efficient hydrolase. At this time, the most effective way to break down plastics involves exposure to UV light together with mechanical disruption caused by waves and winds or grinding on marine rocks and sediments, which eventually breaks larger plastics into smaller pieces of micro- and nanoplastics.[2] This method has its own limitations (including not being environmentally friendly[2]) and microbial degradation of plastics may well be the method of choice in our future endeavors to recycle and/or break down the substantial amount of plastics already polluting almost all of the world’s natural environment.
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 Geyer, R., Jambeck, J. R., & Law, K. L. "Production, use, and fate of all plastics ever made." 2017. Science Advances, 3(7), e1700782.
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 2.13 2.14 2.15 2.16 2.17 2.18 2.19 2.20 2.21 2.22 2.23 2.24 2.25 2.26 2.27 2.28 2.29 2.30 2.31 2.32 2.33 2.34 2.35 2.36 Danso, D., Chow, J., & Streit, W. R. "Plastics: Environmental and Biotechnological Perspectives on Microbial Degradation." 2019b. Applied and Environmental Microbiology, 85(19).
- ↑ Readfearn, G. "'Plastic is literally everywhere': the epidemic attacking Australia's oceans" 2018. The Guardian.
- ↑ Andrei, M. "Plastics are ubiquitous in the deep ocean -- even in the Mariana Trench." 2018. ZME Science.
- ↑ Austin, H. P. et. al. "Characterization and engineering of a plastic-degrading aromatic polyesterase." 2018. Proceedings of the National Academy of Sciences, 115(19), E4350–E4357.
- ↑ Lemonick, S. "Chemistry may have solutions to our plastic trash problem." 2018. Chemical and Engineering News.
- ↑ TOKIWA, Y., & SUZUKI, T. "Hydrolysis of polyesters by lipases." 1917. Nature, 270(5632), 76–78. https://doi.org/10.1038/270076a0
- ↑ 8.00 8.01 8.02 8.03 8.04 8.05 8.06 8.07 8.08 8.09 8.10 8.11 8.12 8.13 8.14 8.15 8.16 8.17 8.18 8.19 Yoshida et al "A bacterium that degrades and assimilates poly(ethylene terephthalate)." 2016. Science, 351(6278), 1196–1199.
- ↑ Wilkinson, G. "Scientists hope new enzyme will 'eat' plastic pollution." 2018. CNN.
- ↑ 10.0 10.1 Tanasupawat, S., Takehana, T., Yoshida, S., Hiraga, K., & Oda, K. "Ideonella sakaiensis sp. nov., isolated from a microbial consortium that degrades poly(ethylene terephthalate)." 2016. International Journal of Systematic and Evolutionary Microbiology, 66(8), 2813–2818.
- ↑ 11.0 11.1 Mathiesen, K. "Could a new plastic-eating bacteria help combat this pollution scourge?" 2016. The Guardian.
- ↑ 12.0 12.1 12.2 12.3 12.4 12.5 Bornscheuer, U. T. "Feeding on plastic." 2016. Science, 351(6278), 1154–1155.
- ↑ Dimarogona, M. et al (2015). "Structural and functional studies of a Fusarium oxysporum cutinase with polyethylene terephthalate modification potential." 2015. Biochimica et Biophysica Acta (BBA) - General Subjects, 1850(11), 2308–2317.
- ↑ 14.0 14.1 Carrington, D. "Scientists accidentally create mutant enzyme that eats plastic bottles." 2018. The Guardian.
- ↑ 15.0 15.1 Müller, R. et al "Enzymatic Degradation of Poly(ethylene terephthalate): Rapid Hydrolyse using a Hydrolase from T. fusca." 2005. Macromolecular Rapid Communications, 26(17), 1400–1405.
- ↑ 16.0 16.1 16.2 16.3 16.4 16.5 16.6 Austin, H. P. et al "Characterization and engineering of a plastic-degrading aromatic polyesterase." 2018b. Proceedings of the National Academy of Sciences, 115(19), E4350–E4357.
- ↑ Kale S.K., Deshmukh, A.G., Dudhare, M.S., & Patil, V.B. "MICROBIAL DEGRADATION OF PLASTIC: A REVIEW." 2015. SEMANTICS SCHOLAR
- ↑ 18.0 18.1 18.2 18.3 18.4 18.5 Espinosa, M. J. C. et al "Toward Biorecycling: Isolation of a Soil Bacterium That Grows on a Polyurethane Oligomer and Monomer." 2020. Frontiers in Microbiology, 11.
- ↑ 19.0 19.1 Restrepo-Flórez, J.-M., Bassi, A., & Thompson, M. R. "Microbial degradation and deterioration of polyethylene – A review." 2014. International Biodeterioration & Biodegradation, 88, 83–90.
- ↑ 20.0 20.1 Cassone, B. J. et al. "Role of the intestinal microbiome in low-density polyethylene degradation by caterpillar larvae of the greater wax moth, Galleria mellonella." 2020. Proceedings of the Royal Society B: Biological Sciences, 287(1922), 20200112.
- ↑ 21.0 21.1 21.2 21.3 21.4 21.5 A. K., Rymowicz, W., & Mirończuk, A. M. "Degradation of plastics and plastic-degrading bacteria in cold marine habitats." 2018. Applied Microbiology and Biotechnology, 102(18), 7669–7678.
- ↑ 22.0 22.1 Ho, B. T., Roberts, T. K., & Lucas, S. "An overview on biodegradation of polystyrene and modified polystyrene: the microbial approach." 2017. Critical Reviews in Biotechnology, 38(2), 308–320.
- ↑ 23.0 23.1 23.2 23.3 23.4 Taniguchi, I. et al. "Biodegradation of PET: Current Status and Application Aspects." 2019. ACS Catalysis, 9(5), 4089–4105.
- ↑ O' Leary, N.D. et al. "Regulation of phenylacetic acid uptake is σ54 dependent in Pseudomonas putidaCA-3." 2011. BMC Microbiol 11, 229.
- ↑ Widyastuti, G. "Genetic Engineered Ideonella Sakaiensis Bacteria: A Solution of the Legendary Plastic Waste Problem." 2018. SSRN Electronic Journal.
Authored for BIOL 238 Microbiology, taught by Joan Slonczewski, 2020, Kenyon College.
