Sindbis Virus: Difference between revisions
| (93 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
{{Uncurated}} | {{Uncurated}} | ||
= | =Classification= | ||
Sindbis virus is a species that occupies human, invertebrate and vertebrate hosts ( | Sindbis virus is a species that occupies human, invertebrate and vertebrate hosts<ref name=one>Schoch, C. L., Ciufo, S., Domrachev, M., Hotton, C. L., Kannan, S., Khovanskaya, R., | ||
Leipe, D., Mcveigh, R., O’Neill, K., Robbertse, B., Sharma, S., Soussov, V., Sullivan, J. P., Sun, L., Turner, S., & Karsch-Mizrachi, I. (2020). <i>Sindbis virus</i>. NCBI Taxonomy Browser. https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?lvl=0&id=11034 | |||
</ref>. | |||
Domain: Viruses | Domain: Viruses | ||
Realm: Riboviria | |||
Kingdom: Orthornavirae | |||
Phylum: Kitrinoviricota | |||
Class: Alsuviricetes | |||
Order: Martellivirales | |||
=6. | Family: Togaviridae | ||
=7. Pathology= | Genus: Alphavirus | ||
= | =Introduction= | ||
Sindbis virus (SINV) is a disease-causing RNA virus transferred from animals to humans that is most often observed in Northern Europe and South Africa<ref name=two>Adouchief, S., Smura, T., Sane, J., Vapalahti, O., & Kurkela, S. (2016, March 15). Sindbis virus as a human pathogen— epidemiology, clinical picture and pathogenesis. <i>Reviews in Medical Virology, 26</i>(1), 221-241. https://www.doi.org/10.1002/rmv.1876</ref>. The clinical infection of SINV is known as Pogosta disease in Finland, Ockelbo disease in Sweden, and Karelian fever in Russia. Symptoms of infection include joint pain, rash, fever, and myalgia; in many patients, these symptoms can continue for months or years and become debilitating<ref name=two/>. | |||
= | |||
It is | After diagnosis, there is no specific treatment available but pain management is typically used to alleviate symptoms through anti-inflammatory drugs<ref name=two/>. Potential cancer therapies have been developed using a viral vector of SINV to introduce specific genomic information into a host cell<ref name=three>Quetglas, J. I., Ruiz-Guillen, M., Aranda, A., Casales, E., Bezunartea, J., & Smerdou, C. (2010). Alphavirus vectors for cancer therapy. <i>Virus Research, 153</i>(2), 179–196. https://doi.org/10.1016/j.virusres.2010.07.027</ref>. However, it is unknown whether SINV vectors are effective in a clinical setting outside of a lab<ref name=two/>. It is also not yet well understood how host cells signal and respond to infection of SINV<ref name=two/>. | ||
Current research is focused on understanding the pathophysiology of SINV so that effective prevention and treatment methods for the virus can be developed<ref name=three/>, as well as expanding the phylogenetic tree of different SINV strains<ref name=four>Strauss, E. G., Rice, C. M., & Strauss, J. H. (1984). Complete nucleotide sequence of the genomic RNA of Sindbis virus. <i>Virology, 133</i>(1), 92-110. https://doi.org/10.1016/0042-6822(84)90428-8</ref>. | |||
=Genome structure= | |||
The genome of SINV is a single-stranded positive sense RNA molecule<ref name=four/>. Among the 44 strains that have been sequenced, the genome ranges from 11-12 kilobases and is additionally capped at the 5' end and polyadenylated at the 3' end<ref name=four/><ref name=five>Sem Ouilibona, R. C., Tchetgna Simo, H. D., Vickos, U., Berthet, N., & Nakoune, E. (2018, June 28). Full-length genome sequence of a Sindbis virus strain isolated from Culex cinereus in 1977 in Bozo, Central African Republic. <i>Genome Announcements, 6</i>(26). https://doi.org/10.1128/genomeA.00455-18</ref><ref name=six>Korhonen, E., Suvanto, M., Uusitalo, R., Faolotto, G., Smura, T., Sane, J., Vapalahti, O., & Huhtamo, E. (2020, October 28). Sindbis virus strains of divergent origin isolated from humans and mosquitoes during a recent outbreak in Finland. <i>Vector-Borne and Zoonotic Diseases, 20</i>(11), 843-849. https://doi.org/10.1089/vbz.2019.2562</ref>. The highest variance between SINV strains at the amino acid level was 22.2% and currently breaks down into 5 genotypic groups: SIN-I (Europe and Africa), SIN-II (Australia), SIN-III (East Asia), SIN-IV (Azerbaijan and China), and SIN-V (New Zealand)<ref name=seven>Lundström, J., Pfeffer, M. (2010, November). Phylogeographic structure and evolutionary history of Sindbis virus. <i>Vector-Borne and Zoonotic Diseases, 10</i>(9), 1-16. https://doi.org/10.1089/vbz.2009.0069</ref>. The viral genome encodes mRNA for five structural proteins and four nonstructural proteins<ref name=two/>. | |||
The small plaque, heat resistant strain of SINV has a genome that is 11,703 nucleotides long. The genome has 59 nucleotides of 5' untranslated regions from the 5’ cap<ref name=four/>. Following the 5’ untranslated region are 7539 nucleotides of open reading frame that encode nonstructural polypeptides (nsPs), nsP1, nsP2, nsP3, and nsP4, which are involved in transcription and translation of the viral genome<ref name=four/>. Two different polyprotein precursors can be produced from this part of the genome, either P123 or P1234<ref name=four/><ref name=eight>Abu Bakar, F., & Ng, L. F. P. (2018). Nonstructural proteins of Alphavirus—potential targets for drug development. <i>Viruses, 10</i>(2), 71. https://doi.org/10.3390/v10020071</ref>. The nonstructural proteins then work in the replication complex to create complementary RNAs in the process of viral genome replication<ref name=eight/><ref name=nine>Spuul, P., Balistreri, G., Hellström, K., Golubtsov, A. V., Jokitalo, E., & Ahola, T. (2011). Assembly of Alphavirus replication complexes from RNA and protein components in a novel trans-replication system in mammalian cells. <i>Journal of Virology, 85</i>(10), 4739-4751. https://doi.org/doi:10.1128/JVI.00085-11</ref>. Following the first open reading frame are 48 untranslated nucleotides located between the sequence encoding the nonstructural proteins and the structural proteins<ref name=four/>. | |||
After the 48 untranslated nucleotides, the next 3735 nucleotides of the open reading frame encode structural proteins which are translated to a polypeptide precursor from the mRNA<ref name=four/>. This precursor is then processed through protein breakdown into the nucleocapsid protein, two integral membrane glycoproteins, and two small peptides (the last two are not found in the mature virus particle). These proteins are named capsid, E1 and E2, and E3 and 6K, respectively<ref name=four/><ref name=ten>Zhang, W., Mukhopadhyay, S., Pletnev, S. V., Baker, T. S., Kuhn, R. J., & Rossmann, M. G. (2002). Placement of the structural proteins in Sindbis virus. <i>Journal of Virology, 76</i>(22), 11645–11658. https://doi.org/10.1128/jvi.76.22.11645-11658.2002</ref>. Found at the end are 322 nucleotides of the 3' untranslated region<ref name=four/>. | |||
=Replication= | |||
SINV uses receptor-mediated endocytosis to enter host cells through clathrin-coated vesicles<ref name=two/><ref name=four/><ref name=five/>. However, recent research has indicated that the genome of SINV may also enter directly through pores formed at the host cell membrane<ref name=two/>. The first step in the replication of the viral genome is the translation of the viral mRNA, allowing the nonstructural proteins to be expressed<ref name=ten/>. The nonstructural proteins then assemble with host factors and become the replication complex<ref name=eight/><ref name=nine/>. The replication complex replicates the viral mRNA and produces the complementary negative sense strand RNAs<ref name=nine/>. Using the negative strand as a template, new positive RNA strands are made by the replication complex and are eventually incorporated into new viral particles<ref name=nine/><ref name=ten/>. | |||
To create new viral particles, the E1 and E2 glycoproteins, two of the structural proteins, get translated into transmembrane proteins of the host cell<ref name=ten/>. After folding, the two proteins form heterodimers and sugars are added via glycosylation in the Golgi apparatus. Following this post-translational modification, the dimer is transported to the plasma membrane of the host cell and undergoes further organization into spikes by assembling with two other dimers to become a trimeric spike<ref name=ten/>. 80 of these spikes assemble to form an icosahedral structure around the virus<ref name=ten/>. | |||
While the viral proteins are being assembled, nucleocapsid cores are formed in the cytoplasm of host cells<ref name=ten/>. The nucleocapsid cores are transported to the plasma membrane of the cell where they interact with the cytoplasmic domains of the E1-E2 trimers. When viral particles are ready to leave the host cell, the host cell membrane starts to bud<ref name=ten/>. In this budding process, the glycoproteins and the plasma membrane form a viral envelope in the aforementioned icosahedral structure. The capsid, E1, and E2 proteins compose the mature virus in combination with a host cell-derived membrane<ref name=eight/><ref name=ten/> (Figure 2). The new particle can then continue to infect other host cells<ref name=ten/>. | |||
Heparan sulfate, a carbohydrate chain expressed on the surface of animal cells, participates in the binding of SINV to host cells<ref name=twelwe>Byrnes, A. P., & Griffin, D. E. (2020, December 22). Binding of Sindbis virus to cell surface heparan sulfate. <i>Journal of Virology, 72</i>(9), 7349-7356. https://doi.org/10.1128/JVI.72.9.7349-7356.1998</ref>. The amount of heparan sulfate present is a crucial factor that determines how strongly the virus can bind to host cells<ref name=twelwe/>. When the virus is bound to heparan sulfate, it can then enter and replicate inside the host cell<ref name=twelwe/>. | |||
=Particle structure= | |||
[[Image:Billede1.jpg|thumb|200px|right|Figure 2 [Image reprinted under Creative Commons License<ref name=thirteen>National Museum of Natural History (2021). <i>Sindbis virus</i>. Encyclopedia of Life. https://eol.org/pages/539944</ref>]. ''Cryo-electron microscopy of the Sindbis virus - liposome complex''.]] | |||
SINV is an enveloped virus, meaning the virus is wrapped in a coat of the host cell’s plasma membrane<ref name=ten/>. This membrane holds 80 spike proteins, each composed of 3 heterodimers, where each heterodimer consists of a pair of E1 and E2 glycoproteins (peptide chains with carbohydrate groups attached)<ref name=ten/>. It is a spherical virus about 70 nanometers in diameter that contains an icosahedral, or 20-sided, capsid (Figure 2). | |||
=Metabolic processes= | |||
SINV does not have its own metabolic processes<ref name=fourteen>Findlay, J. S., & Ulaeto, D. (2015). Semliki Forest virus and Sindbis virus, but not Vaccinia virus, require glycolysis for optimal replication. <i>Journal of General Virology, 96</i>(9), 2693-2696. https://doi.org/10.1099/jgv.0.000226</ref>. SINV is an obligate pathogen that depends on its hosts’ cell machinery to generate energy for replication and survival. When kidney cells from African Green monkeys were infected with SINV following incubation with glycolysis inhibitors, SINV replication was found to be lower than infected cells not incubated with glycolysis inhibitors<ref name=fourteen/>. | |||
SINV also manipulates host cell metabolic processes to favor efficient viral replication<ref name=fifteen>Silva da Costa, L., Pereira da Silva, A. P., Da Poian, A. T., & El-Bacha, T. (2012, April 2). Mitochondrial bioenergetic alterations in mouse neuroblastoma cells infected with Sindbis virus: implications to viral replication and neuronal death. <i>PLoS One, 7</i>(4), e33871. https://doi.org/10.1371/journal.pone.0033871</ref>. After 24 hours of infection with SINV, mouse neuroblastoma cells showed mitochondrial impairment, upregulation of glucose uptake and glycolysis, and decreased cellular ATP content. Although upregulation of glucose uptake and glycolysis generates less ATP than fully-functioning mitochondria, it may be important for other pathways useful in viral replication<ref name=fifteen/><ref name=sixteen>Vastag, L., Koyuncu, E., Grady, S. L., Shenk, T. E., & Rabinowitz, J. D. (2011). Divergent effects of human Cytomegalovirus and Herpes simplex virus-1 on cellular metabolism. <i>PLoS Pathogens, 7</i>(7), e1002124. https://doi.org/10.1371/journal.ppat.1002124</ref>. One pathway SINV relies on heavily is the pentose phosphate pathway (PPP)<ref name=seventeen>Mazzon, M., Castro, C., Thaa, B., Liu, L., Mutso, M., Liu, X., Mahalingam, S., Griffin, J., Marsh, M., & McInerney, G. (2018, January 29). Alphavirus-induced hyperactivation of PI3K/AKT directs proviral metabolic changes. <i>PLoS Pathology 14</i>(1), e1006835. https://doi.org/10.1371/journal.ppat.1006835</ref>. The PPP in the host cell aids in the synthesis of viral nucleic acids and lipids which are important for replication. By upregulating glucose uptake and glycolysis, SINV also upregulates the PPP, thereby generating the energy and resources needed for replication<ref name=seventeen/>. | |||
=Ecology= | |||
[[Image:Visual diagram.png|thumb|300px|right|Figure 3 [Image reprinted under Creative Commons License<ref name=twentyone>Lundström, J. O., Hesson, J. C., Schäfer, M. L., Östmann, Ö., Semmler, T., Bekaert, M., Weidmann, M., Lundkvist, Å., & Pfeffer, M. (2019, August 29). Sindbis virus polyarthritis outbreak signalled by virus prevalence in the mosquito vectors. <i>PLOS Neglected Tropical Diseases, 13</i>(8), e0007702. https://doi.org/10.1371/journal.pntd.0007702</ref>. | |||
''Visual diagram of the primary route for Sindbis virus infection of humans through asymptomatic mammalian and insect carriers. Different species of birds and mosquitoes transmit the virus to each other. Mosquitoes infect humans, but humans cannot transmit Sindbis virus back to mosquitoes because of SINV’s naturally low viral load''.]] | |||
SINV was originally isolated from mosquitoes in Cairo, Egypt, in the 1950s<ref name=two/><ref name=eighteen>Ziegler, U., Fischer, D., Eiden, M., Reuschel, M., Rinder, M., Müller, K., Schwehn, R., Schmidt, V., Groschup, M. H., & Keller, M. (2019, December). Sindbis virus— a wild bird associated zoonotic arbovirus circulates in Germany. <i>Veterinary Microbiology, 239</i>(1), 108453. https://www.doi.org/10.1016/j.vetmic.2019.108453</ref>. Most clinical infections occur during the time of peak activity for mosquitoes, which occurs between April and August<ref name=two/>. Mosquitoes directly transmit the disease to humans, yet birds are the primary amplifying hosts of SINV and spread the virus across geographic areas through migration. Birds are the natural asymptomatic carriers of SINV and transmit the virus to mosquitoes, which can then infect humans. No human-to-human transmission of Sindbis virus has ever been observed<ref name=two/>. | |||
There are many other factors that influence the extent of SINV spread; such as climate (mosquitoes thrive in warmer and wetter conditions), human behavior (such as whether they are outside more often), and herd immunity (if enough people have been infected in the past to develop antibodies against it)<ref name=two/><ref name=eighteen/>. Certain human characteristics have been observed to make one more susceptible to symptomatic infection by SINV — such as female gender, older age, rheumatism, and expression of the human leukocyte antigen allele DRB1*01<ref name=nineteen>Sane, J., Kurkela, S., Lokki, M., Miettinen, A., Helve, T., Vaheri, A., & Vapalahti, O. (2012, August). Clinical Sindbis virus alphavirus infection is associated with HLA-DRB1*01 allele and production of autoantibodies. <i>Clinical Infectious Disease, 55</i>(3), 358-363. https://www.doi.org/10.1093/cid/cis405</ref>. In Finland, environmental variables like warm temperatures, high precipitation, and thick snow coverings from the previous winter are conducive to the spread of SINV<ref name=twenty>Jalava, K., Sane, J., Ollgren, J, Ruuhela, R., Rätti, O., Kurkela, S., Helle, P., Hartonen, S., Pirinen, P., Vapalahti, O., & Kuusi, M. (2012, November 15). Climatic, ecological, and socioeconomic factors as predictors of Sindbis virus infections in Finland. <i>Epidemiology and Infection, 141</i>(9), 1857-1866. https://doi.org/10.1017/S095026881200249X</ref>. These conditions are most prominent in the Central-Eastern region of Finland, where SINV is endemic, and occur most often from July to October. | |||
The largest SINV outbreaks have been reported on a 7 year repeating cycle in addition to annual occurrences in the Central-Eastern region of Finland. One determinant of this timescale is a higher density in black grouse hosts during a given hatch year, followed by the development of infected mosquitoes and subsequent transmission of the virus to humans usually during outdoor activities<ref name=twenty/>. | |||
=Pathology= | |||
Symptom onset for SINV occurs after the incubation period, which is on average about 4 days after infection<ref name=seventeen/><ref name=twentytwo>Lopes Marques, C. D., Ranzolin, A., Cavalcanti, N. G., Luzia, A., Pinto Duarte, B. (2020, August). Arboviruses related with chronic musculoskeletal symptoms. <i>Best Practice & Research Clinical Rheumatology, 34</i>(4), 101502. https://doi.org/10.1016/j.berh.2020.101502</ref>. Compared to other alphaviruses, the length of time that SINV has a viral load in the human blood is much lower, being <103 RNA copies/mL of blood serum in acutely ill patients sick with SINV. The human body starts making antibodies in detectable amounts within the first week of infection<ref name=seventeen/><ref name=twentytwo/>. The symptoms caused by SINV start mild and can become severe depending on the patient. Mild symptoms include skin rashes, malaise, fever, and headache, while progression of illness may include musculoskeletal symptoms such as joint pain in ankles, fingers, wrists and knees, and myalgia which can restrict activities of daily living and limit mobility<ref name=seventeen/><ref name=twentytwo/>. | |||
Heparan sulfate facilitates SINV infection in a host. Heparan sulfate is a carbohydrate chain expressed on the surface of animal cells and binds SINV<ref name=twelwe/>. The amount of sulfate present is a crucial factor that determines how strongly the virus can bind to host cells<ref name=twelwe/>. The cellular antiviral endoribonuclease RNase L cleaves the viral genome to produce small RNAs in response to SINV infection<ref name=twentythree>Girardi, E., Chane-Woon-Ming, B., Messmer, M., Kaukinen, P., & Pfeffer, S. (2013, December 31). Identification of RNase L-dependent, 3’-end-modified, viral small RNAs in Sindbis virus-infected mammalian cells. <i>mBio, 4</i>(6). https://doi.org/10.1128/mBio.00698-13</ref>. These small viral RNAs are modified and stabilized, which may indicate their use in cell signaling for host immune response and degradation<ref name=twentythree/>. | |||
Mutations in capping activity affect SINV viral load. nsP1 capping mutants can be used to observe the effect of noncapped genomic RNAs (ncgRNA) on alphaviral load<ref name=twentyfour>LaPointe, A. T., Douglas Landers, V., Westcott, C. E., & Sokoloski, K. J. (2020, December 1). Production of noncapped genomic RNAs is critical to Sindbis virus disease and pathogenicity. <i>mBio, 11</i>(6). https://doi.org//10.1128/mBio.02675-20</ref>. D355A and N376A nsP1 mutations, which function to increase and decrease capping activity respectively, were inserted into the AR86 strain of Sindbis virus. Increasing capping efficiency with the D355A point mutation decreased the production of ncgRNA, reduced SINV viral load, and reduced mortality and morbidity in that cohort of infected mice<ref name=twentyfour/>. Mutating caps for uncapped genomic RNA) provide a route of investigation for the development of vaccines and treatments for Sindbis virus<ref name=twentyfour/>. | |||
It is not well understood why certain people are more susceptible to severe infection from SINV compared to others; however, several genetic factors are associated with patient outcomes. There is a link between the gene family for the human leukocyte antigen complex, which helps a person’s immune system distinguish their own proteins from proteins made by viruses or bacteria, and clinical infection of SINV<ref name=eighteen/>. The HLA-DRB1*01 allele was notably higher in patients with SINV infection compared to the general population and was particularly high in patients who continued to have joint pain three years after infection. These results show similarities to the genetic factors which predispose people to acquire autoimmune diseases, which could be an important avenue to find a treatment for SINV using the same strategies<ref name=eighteen/>. | |||
Living cells can become infected by SINV even without endocytosis and membrane fusion: SINV particles have emptied themselves of their RNA genome and inserted it into host cells at temperatures as low as 4℃/39.2℉, where these processes are inhibited<ref name=twentyfive>Vancini, R., Wang, G., Ferreira, D., Hernandez, R., & Brown, D. T. (2013). Alphavirus genome delivery occurs directly at the plasma membrane in a time- and temperature-dependent process. <i>Journal of Virology, 87</i>(8), 4352. https://www.doi.org/10.1128/JVI.03412-12</ref>. However, while host infection is possible in these conditions, optimal conditions are at 37℃/98.6℉— when endocytosis and membrane fusion are uninhibited. | |||
Diagnosis of infection due to Sindbis virus is based on an enzyme immunoassay for detection of SINV-specific antibodies, a test that only a limited number of labs can run<ref name=two/>. This is because the viral load of SINV is so low, averaging about 103 RNA copies per mL in acutely ill patients, that polymerase chain reaction (PCR) is an unrealistic diagnostic test<ref name=three/>. | |||
=Current Research= | |||
One avenue of current research investigates the uses of SINV vectors in medicine. Oncolytic virus therapy uses virus vectors to infect and lyse tumor cells<ref name=twentysix>Yu, M., Scherwitzl, I., Opp, S., Tsirigos, A., & Meruelo, D. (2019, July 15). Molecular and metabolic pathways mediating curative treatment of a non-Hodgkin B cell lymphoma by Sindbis viral vectors and anti-4-1BB monoclonal antibody. <i>Journal for Immunotherapy of Cancer, 185</i>(7), 1-17. https://doi.org/10.1186/s40425-019-0664-3</ref>. In mice with B-cell lymphoma, the use of a Sindbis vector in combination with chemotherapy eliminated a tumor entirely in one mouse. In all other mice, it produced inflammation, triggering an anti-tumor immune response<ref name=twentysix/>. Compared to current therapies, oncolytic virus therapies have a much shorter processing time and may be more effective than chemotherapy alone. | |||
Treatment with an agonist (ACEA) versus an antagonist/inverse agonist (AM-251) was investigated to see how it affects cellular metabolism and SINV viral load when interacting with the cannabinoid receptor 1, called CB1<ref name=twentyseven>Rodriguez, J. L., Lopez, J. A., & Steel, J. J. (2021). Involvement of the endocannabinoid system in the inhibition of Sindbis virus replication: a preliminary study. <i>Journal of Cannabis Research, 3</i>(1), 10. https://doi.org/10.1186/s42238-021-00068-y</ref>. Treatment with AM-251 reduced SINV viral replication in human liver cells, whereas treatment with ACEA facilitated SINV replication. Thus, when AM-251 blocks the CB1 signaling pathway, SINV viral replication is significantly decreased<ref name=twentyseven/>. These findings are useful for continued research on treatment for SINV infection. | |||
Another route of current research investigates the production of an interferon-stimulated host factor called zinc finger antiviral protein (ZAP), which is used as part of the immune response to inhibit SINV replication in the host<ref name=twentyeight>Wang, X., Li, M. M. H., Zhao, J., Li, S., MacDonald, M. R., Rice, C. M., Gao, X., & Gao, G. (2016, August 31). Sindbis virus can exploit a host antiviral protein to evade immune surveillance. <i>Journal of Virology, 22</i>(15), 10247-10258. https://www.doi.org/10.1128/JVI.01487-16</ref>. ZAP works by binding to and inhibiting the translation of target mRNA, as well as by promoting its degradation. Cell cultures from ZAP-deficient mice had higher mortality rates compared to wild-type mice<ref name=twentyeight/>. Additionally, hosts whose interferon activity were blocked became more vulnerable to SINV infection; this is because the virus utilized ZAP to limit its replication and avoid detection from the mice’ immune system — thus allowing SINV to reach the host’s central nervous system<ref name=twentyeight/>. | |||
=Authorship Statement= | |||
Classification: Emily Gant | |||
Introduction: Nicole Amodio, Heather Cook | |||
Genome Structure & Particle Structure: HC, Vianne Nielsen | |||
Metabolic Processes: EG, Matthew Yu | |||
Ecology: HC, MY | |||
Pathology: NA, HC, EG, VN | |||
Current Research: NA, HC, EG | |||
Edits: NA, HC, EG, VN, MY | |||
Edited by Emily Gant & Vianne Nielsen, students of [mailto:jmbhat@bu.edu Jennifer Bhatnagar] for [http://www.bu.edu/academics/cas/courses/cas-bi-311/ BI 311 General Microbiology], 2021, [http://www.bu.edu/ Boston University]. | |||
=References= | |||
Latest revision as of 16:04, 14 January 2022
Classification
Sindbis virus is a species that occupies human, invertebrate and vertebrate hosts[1].
Domain: Viruses
Realm: Riboviria
Kingdom: Orthornavirae
Phylum: Kitrinoviricota
Class: Alsuviricetes
Order: Martellivirales
Family: Togaviridae
Genus: Alphavirus
Introduction
Sindbis virus (SINV) is a disease-causing RNA virus transferred from animals to humans that is most often observed in Northern Europe and South Africa[2]. The clinical infection of SINV is known as Pogosta disease in Finland, Ockelbo disease in Sweden, and Karelian fever in Russia. Symptoms of infection include joint pain, rash, fever, and myalgia; in many patients, these symptoms can continue for months or years and become debilitating[2].
After diagnosis, there is no specific treatment available but pain management is typically used to alleviate symptoms through anti-inflammatory drugs[2]. Potential cancer therapies have been developed using a viral vector of SINV to introduce specific genomic information into a host cell[3]. However, it is unknown whether SINV vectors are effective in a clinical setting outside of a lab[2]. It is also not yet well understood how host cells signal and respond to infection of SINV[2].
Current research is focused on understanding the pathophysiology of SINV so that effective prevention and treatment methods for the virus can be developed[3], as well as expanding the phylogenetic tree of different SINV strains[4].
Genome structure
The genome of SINV is a single-stranded positive sense RNA molecule[4]. Among the 44 strains that have been sequenced, the genome ranges from 11-12 kilobases and is additionally capped at the 5' end and polyadenylated at the 3' end[4][5][6]. The highest variance between SINV strains at the amino acid level was 22.2% and currently breaks down into 5 genotypic groups: SIN-I (Europe and Africa), SIN-II (Australia), SIN-III (East Asia), SIN-IV (Azerbaijan and China), and SIN-V (New Zealand)[7]. The viral genome encodes mRNA for five structural proteins and four nonstructural proteins[2].
The small plaque, heat resistant strain of SINV has a genome that is 11,703 nucleotides long. The genome has 59 nucleotides of 5' untranslated regions from the 5’ cap[4]. Following the 5’ untranslated region are 7539 nucleotides of open reading frame that encode nonstructural polypeptides (nsPs), nsP1, nsP2, nsP3, and nsP4, which are involved in transcription and translation of the viral genome[4]. Two different polyprotein precursors can be produced from this part of the genome, either P123 or P1234[4][8]. The nonstructural proteins then work in the replication complex to create complementary RNAs in the process of viral genome replication[8][9]. Following the first open reading frame are 48 untranslated nucleotides located between the sequence encoding the nonstructural proteins and the structural proteins[4].
After the 48 untranslated nucleotides, the next 3735 nucleotides of the open reading frame encode structural proteins which are translated to a polypeptide precursor from the mRNA[4]. This precursor is then processed through protein breakdown into the nucleocapsid protein, two integral membrane glycoproteins, and two small peptides (the last two are not found in the mature virus particle). These proteins are named capsid, E1 and E2, and E3 and 6K, respectively[4][10]. Found at the end are 322 nucleotides of the 3' untranslated region[4].
Replication
SINV uses receptor-mediated endocytosis to enter host cells through clathrin-coated vesicles[2][4][5]. However, recent research has indicated that the genome of SINV may also enter directly through pores formed at the host cell membrane[2]. The first step in the replication of the viral genome is the translation of the viral mRNA, allowing the nonstructural proteins to be expressed[10]. The nonstructural proteins then assemble with host factors and become the replication complex[8][9]. The replication complex replicates the viral mRNA and produces the complementary negative sense strand RNAs[9]. Using the negative strand as a template, new positive RNA strands are made by the replication complex and are eventually incorporated into new viral particles[9][10].
To create new viral particles, the E1 and E2 glycoproteins, two of the structural proteins, get translated into transmembrane proteins of the host cell[10]. After folding, the two proteins form heterodimers and sugars are added via glycosylation in the Golgi apparatus. Following this post-translational modification, the dimer is transported to the plasma membrane of the host cell and undergoes further organization into spikes by assembling with two other dimers to become a trimeric spike[10]. 80 of these spikes assemble to form an icosahedral structure around the virus[10].
While the viral proteins are being assembled, nucleocapsid cores are formed in the cytoplasm of host cells[10]. The nucleocapsid cores are transported to the plasma membrane of the cell where they interact with the cytoplasmic domains of the E1-E2 trimers. When viral particles are ready to leave the host cell, the host cell membrane starts to bud[10]. In this budding process, the glycoproteins and the plasma membrane form a viral envelope in the aforementioned icosahedral structure. The capsid, E1, and E2 proteins compose the mature virus in combination with a host cell-derived membrane[8][10] (Figure 2). The new particle can then continue to infect other host cells[10].
Heparan sulfate, a carbohydrate chain expressed on the surface of animal cells, participates in the binding of SINV to host cells[11]. The amount of heparan sulfate present is a crucial factor that determines how strongly the virus can bind to host cells[11]. When the virus is bound to heparan sulfate, it can then enter and replicate inside the host cell[11].
Particle structure

SINV is an enveloped virus, meaning the virus is wrapped in a coat of the host cell’s plasma membrane[10]. This membrane holds 80 spike proteins, each composed of 3 heterodimers, where each heterodimer consists of a pair of E1 and E2 glycoproteins (peptide chains with carbohydrate groups attached)[10]. It is a spherical virus about 70 nanometers in diameter that contains an icosahedral, or 20-sided, capsid (Figure 2).
Metabolic processes
SINV does not have its own metabolic processes[13]. SINV is an obligate pathogen that depends on its hosts’ cell machinery to generate energy for replication and survival. When kidney cells from African Green monkeys were infected with SINV following incubation with glycolysis inhibitors, SINV replication was found to be lower than infected cells not incubated with glycolysis inhibitors[13].
SINV also manipulates host cell metabolic processes to favor efficient viral replication[14]. After 24 hours of infection with SINV, mouse neuroblastoma cells showed mitochondrial impairment, upregulation of glucose uptake and glycolysis, and decreased cellular ATP content. Although upregulation of glucose uptake and glycolysis generates less ATP than fully-functioning mitochondria, it may be important for other pathways useful in viral replication[14][15]. One pathway SINV relies on heavily is the pentose phosphate pathway (PPP)[16]. The PPP in the host cell aids in the synthesis of viral nucleic acids and lipids which are important for replication. By upregulating glucose uptake and glycolysis, SINV also upregulates the PPP, thereby generating the energy and resources needed for replication[16].
Ecology
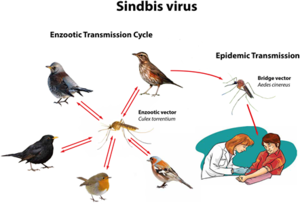
SINV was originally isolated from mosquitoes in Cairo, Egypt, in the 1950s[2][18]. Most clinical infections occur during the time of peak activity for mosquitoes, which occurs between April and August[2]. Mosquitoes directly transmit the disease to humans, yet birds are the primary amplifying hosts of SINV and spread the virus across geographic areas through migration. Birds are the natural asymptomatic carriers of SINV and transmit the virus to mosquitoes, which can then infect humans. No human-to-human transmission of Sindbis virus has ever been observed[2].
There are many other factors that influence the extent of SINV spread; such as climate (mosquitoes thrive in warmer and wetter conditions), human behavior (such as whether they are outside more often), and herd immunity (if enough people have been infected in the past to develop antibodies against it)[2][18]. Certain human characteristics have been observed to make one more susceptible to symptomatic infection by SINV — such as female gender, older age, rheumatism, and expression of the human leukocyte antigen allele DRB1*01[19]. In Finland, environmental variables like warm temperatures, high precipitation, and thick snow coverings from the previous winter are conducive to the spread of SINV[20]. These conditions are most prominent in the Central-Eastern region of Finland, where SINV is endemic, and occur most often from July to October.
The largest SINV outbreaks have been reported on a 7 year repeating cycle in addition to annual occurrences in the Central-Eastern region of Finland. One determinant of this timescale is a higher density in black grouse hosts during a given hatch year, followed by the development of infected mosquitoes and subsequent transmission of the virus to humans usually during outdoor activities[20].
Pathology
Symptom onset for SINV occurs after the incubation period, which is on average about 4 days after infection[16][21]. Compared to other alphaviruses, the length of time that SINV has a viral load in the human blood is much lower, being <103 RNA copies/mL of blood serum in acutely ill patients sick with SINV. The human body starts making antibodies in detectable amounts within the first week of infection[16][21]. The symptoms caused by SINV start mild and can become severe depending on the patient. Mild symptoms include skin rashes, malaise, fever, and headache, while progression of illness may include musculoskeletal symptoms such as joint pain in ankles, fingers, wrists and knees, and myalgia which can restrict activities of daily living and limit mobility[16][21].
Heparan sulfate facilitates SINV infection in a host. Heparan sulfate is a carbohydrate chain expressed on the surface of animal cells and binds SINV[11]. The amount of sulfate present is a crucial factor that determines how strongly the virus can bind to host cells[11]. The cellular antiviral endoribonuclease RNase L cleaves the viral genome to produce small RNAs in response to SINV infection[22]. These small viral RNAs are modified and stabilized, which may indicate their use in cell signaling for host immune response and degradation[22].
Mutations in capping activity affect SINV viral load. nsP1 capping mutants can be used to observe the effect of noncapped genomic RNAs (ncgRNA) on alphaviral load[23]. D355A and N376A nsP1 mutations, which function to increase and decrease capping activity respectively, were inserted into the AR86 strain of Sindbis virus. Increasing capping efficiency with the D355A point mutation decreased the production of ncgRNA, reduced SINV viral load, and reduced mortality and morbidity in that cohort of infected mice[23]. Mutating caps for uncapped genomic RNA) provide a route of investigation for the development of vaccines and treatments for Sindbis virus[23].
It is not well understood why certain people are more susceptible to severe infection from SINV compared to others; however, several genetic factors are associated with patient outcomes. There is a link between the gene family for the human leukocyte antigen complex, which helps a person’s immune system distinguish their own proteins from proteins made by viruses or bacteria, and clinical infection of SINV[18]. The HLA-DRB1*01 allele was notably higher in patients with SINV infection compared to the general population and was particularly high in patients who continued to have joint pain three years after infection. These results show similarities to the genetic factors which predispose people to acquire autoimmune diseases, which could be an important avenue to find a treatment for SINV using the same strategies[18].
Living cells can become infected by SINV even without endocytosis and membrane fusion: SINV particles have emptied themselves of their RNA genome and inserted it into host cells at temperatures as low as 4℃/39.2℉, where these processes are inhibited[24]. However, while host infection is possible in these conditions, optimal conditions are at 37℃/98.6℉— when endocytosis and membrane fusion are uninhibited.
Diagnosis of infection due to Sindbis virus is based on an enzyme immunoassay for detection of SINV-specific antibodies, a test that only a limited number of labs can run[2]. This is because the viral load of SINV is so low, averaging about 103 RNA copies per mL in acutely ill patients, that polymerase chain reaction (PCR) is an unrealistic diagnostic test[3].
Current Research
One avenue of current research investigates the uses of SINV vectors in medicine. Oncolytic virus therapy uses virus vectors to infect and lyse tumor cells[25]. In mice with B-cell lymphoma, the use of a Sindbis vector in combination with chemotherapy eliminated a tumor entirely in one mouse. In all other mice, it produced inflammation, triggering an anti-tumor immune response[25]. Compared to current therapies, oncolytic virus therapies have a much shorter processing time and may be more effective than chemotherapy alone.
Treatment with an agonist (ACEA) versus an antagonist/inverse agonist (AM-251) was investigated to see how it affects cellular metabolism and SINV viral load when interacting with the cannabinoid receptor 1, called CB1[26]. Treatment with AM-251 reduced SINV viral replication in human liver cells, whereas treatment with ACEA facilitated SINV replication. Thus, when AM-251 blocks the CB1 signaling pathway, SINV viral replication is significantly decreased[26]. These findings are useful for continued research on treatment for SINV infection.
Another route of current research investigates the production of an interferon-stimulated host factor called zinc finger antiviral protein (ZAP), which is used as part of the immune response to inhibit SINV replication in the host[27]. ZAP works by binding to and inhibiting the translation of target mRNA, as well as by promoting its degradation. Cell cultures from ZAP-deficient mice had higher mortality rates compared to wild-type mice[27]. Additionally, hosts whose interferon activity were blocked became more vulnerable to SINV infection; this is because the virus utilized ZAP to limit its replication and avoid detection from the mice’ immune system — thus allowing SINV to reach the host’s central nervous system[27].
Authorship Statement
Classification: Emily Gant
Introduction: Nicole Amodio, Heather Cook
Genome Structure & Particle Structure: HC, Vianne Nielsen
Metabolic Processes: EG, Matthew Yu
Ecology: HC, MY
Pathology: NA, HC, EG, VN
Current Research: NA, HC, EG
Edits: NA, HC, EG, VN, MY
Edited by Emily Gant & Vianne Nielsen, students of Jennifer Bhatnagar for BI 311 General Microbiology, 2021, Boston University.
References
- ↑ Schoch, C. L., Ciufo, S., Domrachev, M., Hotton, C. L., Kannan, S., Khovanskaya, R., Leipe, D., Mcveigh, R., O’Neill, K., Robbertse, B., Sharma, S., Soussov, V., Sullivan, J. P., Sun, L., Turner, S., & Karsch-Mizrachi, I. (2020). Sindbis virus. NCBI Taxonomy Browser. https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?lvl=0&id=11034
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 Adouchief, S., Smura, T., Sane, J., Vapalahti, O., & Kurkela, S. (2016, March 15). Sindbis virus as a human pathogen— epidemiology, clinical picture and pathogenesis. Reviews in Medical Virology, 26(1), 221-241. https://www.doi.org/10.1002/rmv.1876
- ↑ 3.0 3.1 3.2 Quetglas, J. I., Ruiz-Guillen, M., Aranda, A., Casales, E., Bezunartea, J., & Smerdou, C. (2010). Alphavirus vectors for cancer therapy. Virus Research, 153(2), 179–196. https://doi.org/10.1016/j.virusres.2010.07.027
- ↑ 4.00 4.01 4.02 4.03 4.04 4.05 4.06 4.07 4.08 4.09 4.10 Strauss, E. G., Rice, C. M., & Strauss, J. H. (1984). Complete nucleotide sequence of the genomic RNA of Sindbis virus. Virology, 133(1), 92-110. https://doi.org/10.1016/0042-6822(84)90428-8
- ↑ 5.0 5.1 Sem Ouilibona, R. C., Tchetgna Simo, H. D., Vickos, U., Berthet, N., & Nakoune, E. (2018, June 28). Full-length genome sequence of a Sindbis virus strain isolated from Culex cinereus in 1977 in Bozo, Central African Republic. Genome Announcements, 6(26). https://doi.org/10.1128/genomeA.00455-18
- ↑ Korhonen, E., Suvanto, M., Uusitalo, R., Faolotto, G., Smura, T., Sane, J., Vapalahti, O., & Huhtamo, E. (2020, October 28). Sindbis virus strains of divergent origin isolated from humans and mosquitoes during a recent outbreak in Finland. Vector-Borne and Zoonotic Diseases, 20(11), 843-849. https://doi.org/10.1089/vbz.2019.2562
- ↑ Lundström, J., Pfeffer, M. (2010, November). Phylogeographic structure and evolutionary history of Sindbis virus. Vector-Borne and Zoonotic Diseases, 10(9), 1-16. https://doi.org/10.1089/vbz.2009.0069
- ↑ 8.0 8.1 8.2 8.3 Abu Bakar, F., & Ng, L. F. P. (2018). Nonstructural proteins of Alphavirus—potential targets for drug development. Viruses, 10(2), 71. https://doi.org/10.3390/v10020071
- ↑ 9.0 9.1 9.2 9.3 Spuul, P., Balistreri, G., Hellström, K., Golubtsov, A. V., Jokitalo, E., & Ahola, T. (2011). Assembly of Alphavirus replication complexes from RNA and protein components in a novel trans-replication system in mammalian cells. Journal of Virology, 85(10), 4739-4751. https://doi.org/doi:10.1128/JVI.00085-11
- ↑ 10.00 10.01 10.02 10.03 10.04 10.05 10.06 10.07 10.08 10.09 10.10 10.11 Zhang, W., Mukhopadhyay, S., Pletnev, S. V., Baker, T. S., Kuhn, R. J., & Rossmann, M. G. (2002). Placement of the structural proteins in Sindbis virus. Journal of Virology, 76(22), 11645–11658. https://doi.org/10.1128/jvi.76.22.11645-11658.2002
- ↑ 11.0 11.1 11.2 11.3 11.4 Byrnes, A. P., & Griffin, D. E. (2020, December 22). Binding of Sindbis virus to cell surface heparan sulfate. Journal of Virology, 72(9), 7349-7356. https://doi.org/10.1128/JVI.72.9.7349-7356.1998
- ↑ National Museum of Natural History (2021). Sindbis virus. Encyclopedia of Life. https://eol.org/pages/539944
- ↑ 13.0 13.1 Findlay, J. S., & Ulaeto, D. (2015). Semliki Forest virus and Sindbis virus, but not Vaccinia virus, require glycolysis for optimal replication. Journal of General Virology, 96(9), 2693-2696. https://doi.org/10.1099/jgv.0.000226
- ↑ 14.0 14.1 Silva da Costa, L., Pereira da Silva, A. P., Da Poian, A. T., & El-Bacha, T. (2012, April 2). Mitochondrial bioenergetic alterations in mouse neuroblastoma cells infected with Sindbis virus: implications to viral replication and neuronal death. PLoS One, 7(4), e33871. https://doi.org/10.1371/journal.pone.0033871
- ↑ Vastag, L., Koyuncu, E., Grady, S. L., Shenk, T. E., & Rabinowitz, J. D. (2011). Divergent effects of human Cytomegalovirus and Herpes simplex virus-1 on cellular metabolism. PLoS Pathogens, 7(7), e1002124. https://doi.org/10.1371/journal.ppat.1002124
- ↑ 16.0 16.1 16.2 16.3 16.4 Mazzon, M., Castro, C., Thaa, B., Liu, L., Mutso, M., Liu, X., Mahalingam, S., Griffin, J., Marsh, M., & McInerney, G. (2018, January 29). Alphavirus-induced hyperactivation of PI3K/AKT directs proviral metabolic changes. PLoS Pathology 14(1), e1006835. https://doi.org/10.1371/journal.ppat.1006835
- ↑ Lundström, J. O., Hesson, J. C., Schäfer, M. L., Östmann, Ö., Semmler, T., Bekaert, M., Weidmann, M., Lundkvist, Å., & Pfeffer, M. (2019, August 29). Sindbis virus polyarthritis outbreak signalled by virus prevalence in the mosquito vectors. PLOS Neglected Tropical Diseases, 13(8), e0007702. https://doi.org/10.1371/journal.pntd.0007702
- ↑ 18.0 18.1 18.2 18.3 Ziegler, U., Fischer, D., Eiden, M., Reuschel, M., Rinder, M., Müller, K., Schwehn, R., Schmidt, V., Groschup, M. H., & Keller, M. (2019, December). Sindbis virus— a wild bird associated zoonotic arbovirus circulates in Germany. Veterinary Microbiology, 239(1), 108453. https://www.doi.org/10.1016/j.vetmic.2019.108453
- ↑ Sane, J., Kurkela, S., Lokki, M., Miettinen, A., Helve, T., Vaheri, A., & Vapalahti, O. (2012, August). Clinical Sindbis virus alphavirus infection is associated with HLA-DRB1*01 allele and production of autoantibodies. Clinical Infectious Disease, 55(3), 358-363. https://www.doi.org/10.1093/cid/cis405
- ↑ 20.0 20.1 Jalava, K., Sane, J., Ollgren, J, Ruuhela, R., Rätti, O., Kurkela, S., Helle, P., Hartonen, S., Pirinen, P., Vapalahti, O., & Kuusi, M. (2012, November 15). Climatic, ecological, and socioeconomic factors as predictors of Sindbis virus infections in Finland. Epidemiology and Infection, 141(9), 1857-1866. https://doi.org/10.1017/S095026881200249X
- ↑ 21.0 21.1 21.2 Lopes Marques, C. D., Ranzolin, A., Cavalcanti, N. G., Luzia, A., Pinto Duarte, B. (2020, August). Arboviruses related with chronic musculoskeletal symptoms. Best Practice & Research Clinical Rheumatology, 34(4), 101502. https://doi.org/10.1016/j.berh.2020.101502
- ↑ 22.0 22.1 Girardi, E., Chane-Woon-Ming, B., Messmer, M., Kaukinen, P., & Pfeffer, S. (2013, December 31). Identification of RNase L-dependent, 3’-end-modified, viral small RNAs in Sindbis virus-infected mammalian cells. mBio, 4(6). https://doi.org/10.1128/mBio.00698-13
- ↑ 23.0 23.1 23.2 LaPointe, A. T., Douglas Landers, V., Westcott, C. E., & Sokoloski, K. J. (2020, December 1). Production of noncapped genomic RNAs is critical to Sindbis virus disease and pathogenicity. mBio, 11(6). https://doi.org//10.1128/mBio.02675-20
- ↑ Vancini, R., Wang, G., Ferreira, D., Hernandez, R., & Brown, D. T. (2013). Alphavirus genome delivery occurs directly at the plasma membrane in a time- and temperature-dependent process. Journal of Virology, 87(8), 4352. https://www.doi.org/10.1128/JVI.03412-12
- ↑ 25.0 25.1 Yu, M., Scherwitzl, I., Opp, S., Tsirigos, A., & Meruelo, D. (2019, July 15). Molecular and metabolic pathways mediating curative treatment of a non-Hodgkin B cell lymphoma by Sindbis viral vectors and anti-4-1BB monoclonal antibody. Journal for Immunotherapy of Cancer, 185(7), 1-17. https://doi.org/10.1186/s40425-019-0664-3
- ↑ 26.0 26.1 Rodriguez, J. L., Lopez, J. A., & Steel, J. J. (2021). Involvement of the endocannabinoid system in the inhibition of Sindbis virus replication: a preliminary study. Journal of Cannabis Research, 3(1), 10. https://doi.org/10.1186/s42238-021-00068-y
- ↑ 27.0 27.1 27.2 Wang, X., Li, M. M. H., Zhao, J., Li, S., MacDonald, M. R., Rice, C. M., Gao, X., & Gao, G. (2016, August 31). Sindbis virus can exploit a host antiviral protein to evade immune surveillance. Journal of Virology, 22(15), 10247-10258. https://www.doi.org/10.1128/JVI.01487-16
