The Gut Microbiome and Parkinson's Disease: Difference between revisions
| Line 17: | Line 17: | ||
</br> | </br> | ||
===Parkinson's Disease Phenotypes=== | ===Parkinson's Disease Phenotypes=== | ||
One research team attempted to connect abnormal gut microbiota with common Parkinson's Disease phenotypes (tremor, abnormal gait, poor balance). From fecal samples, they discovered that the mean abundance of Prevotellaceae was 77.6% lower in Parkinson's Disease patients than in healthy controls. By applying generalized linear modeling, the team demonstrated that this difference could not be explained by differences in constipation levels, comorbidities, or medications. Interestingly, their models revealed that increased levels of Ruminococcaceae in Parkinson's Disease patients could be explained by the decrease in Prevotellaceae; that is, Ruminococcaceae increased <i>because</i> Prevotellaceae decreased.<ref name=Scheperjans>Scheperjans, Filip, Velma Aho, Pedro A. B. Pereira, Kaisa Koskinen, Lars Paulin, Eero Pekkonen, Elena Haapaniemi, Seppo Kaakkola, Johanna Eerola-Rautio, Marjatta Pohja, Esko Kinnunen, Kari Murros, Petri Auvinen. (2014). "Gut microbiota are related to Parkinson's disease and clinical phenotype." <i>Movement Disorders</i>, vol. 30(3), 350-358. https://doi.org/10.1002/mds.26069</ref> | One research team attempted to connect abnormal gut microbiota with common Parkinson's Disease phenotypes (tremor, abnormal gait, poor balance). From fecal samples, they discovered that the mean abundance of Prevotellaceae was 77.6% lower in Parkinson's Disease patients than in healthy controls. By applying generalized linear modeling, the team demonstrated that this difference could not be explained by differences in constipation levels, comorbidities, or medications. Interestingly, their models revealed that increased levels of Ruminococcaceae in Parkinson's Disease patients could be explained by the decrease in Prevotellaceae; that is, Ruminococcaceae increased <i>because</i> Prevotellaceae decreased.<ref name=Scheperjans>Scheperjans, Filip, Velma Aho, Pedro A. B. Pereira, Kaisa Koskinen, Lars Paulin, Eero Pekkonen, Elena Haapaniemi, Seppo Kaakkola, Johanna Eerola-Rautio, Marjatta Pohja, Esko Kinnunen, Kari Murros, Petri Auvinen. (2014). "Gut microbiota are related to Parkinson's disease and clinical phenotype." <i>Movement Disorders</i>, vol. 30(3), 350-358. https://doi.org/10.1002/mds.26069</ref>When the researchers attempted to link gut microbiota with phenotype, they discovered that Enterobacteriaceae were more abundant in patients who dominantly displayed abnormal gait/poor balance than patients who dominantly displayed tremors. They speculated that increased levels of Enterobacteriaceae may explain why patients who do not express the tremor phenotype have faster disease progression and worse prognosis.<ref name=Scheperjans/> | ||
</br> | </br> | ||
Another team discovered that Parkinson's Disease patients with higher abundance of Christensenellaceae had worse nonmotor symptoms. Increased Lactobacillaceae and decreased Lachnospiraceae also correlated with worse cases of intellectual impairment, nonmotor symptoms, gait instability, and posture problems.<ref name=Barichella/> | |||
==Interactions with L-Dopa== | ==Interactions with L-Dopa== | ||
Revision as of 16:25, 5 April 2022
by Rebecca Hölzel
Abnormal Gut Microbiome
The conditions that contribute to the development of Parkinson's Disease are likely very complex. A growing body of research has indicated, however, that the gut microbiome may play a key role in disease development and progression. Parkinson's Disease is commonly associated with abnormal gut microbiomes, including increased and decreased counts of bacteria normally found in healthy individuals.[2] However, some researchers remain skeptical that the gut microbiome influences Parkinson's Disease, believing instead that the disease is what causes the abnormal gut bacteria, and not that the abnormal gut bacteria cause the disease.[3]
General Abnormalities
One research team examined the relationship between gut microbiota and Parkinson's Disease by recruiting newly diagnosed and unmedicated Parkinson's disease patients for study. Using antibiotics or having other gut-related diseases constituted grounds for expulsion. Fecal samples were collected from all patients and used to extract bacterial DNA, which provided measures of bacterial diversity and relative abundance of bacterial genera. They discovered that levels of Verrucomicrobia, Verrucomicrobiaceae, and Akkermansia were twofold higher in Parkinson's Disease patients than healthy controls. Levels of Proteobacteria, Enterobacteriaceae, Christensenellaceae, Lactobacillaceae, Coriobacteriaceae, Bifidobacteriaceae, and Parabacteroides also increased in Parkinson's Disease patients, with Roseburia showing a considerable decrease. A significant increase was also seen for Oscillospira and a significant decrease was seen for Ruminococcus.[4]
Gut Inflammation
Intestinal inflammation, common to Parkinson's Disease, has been linked to abnormal gut microbiota through characterization of bacteria as either pro-inflammatory or anti-inflammatory. It has been found that healthy individuals possess far more colonic mucosal bacteria from Coprobacillaceae, Dorea, and Faecalibacterium than Parkinson's Disease patients. Notably, Faecalibacterium have anti-inflammatory properties. Parkinson's Disease patients also have more pro-inflammatory bacteria, including bacteria from the family Oxalobacteraceae. Fecal samples have also shown higher levels of pro-inflammatory bacteria, including Akkermansia, Oscillospira, and Bacteroides. Levels of pro-inflammatory and anti-inflammatory bacteria correlate to Parkinson's Disease duration.[1]
Parkinson's Disease Phenotypes
One research team attempted to connect abnormal gut microbiota with common Parkinson's Disease phenotypes (tremor, abnormal gait, poor balance). From fecal samples, they discovered that the mean abundance of Prevotellaceae was 77.6% lower in Parkinson's Disease patients than in healthy controls. By applying generalized linear modeling, the team demonstrated that this difference could not be explained by differences in constipation levels, comorbidities, or medications. Interestingly, their models revealed that increased levels of Ruminococcaceae in Parkinson's Disease patients could be explained by the decrease in Prevotellaceae; that is, Ruminococcaceae increased because Prevotellaceae decreased.[5]When the researchers attempted to link gut microbiota with phenotype, they discovered that Enterobacteriaceae were more abundant in patients who dominantly displayed abnormal gait/poor balance than patients who dominantly displayed tremors. They speculated that increased levels of Enterobacteriaceae may explain why patients who do not express the tremor phenotype have faster disease progression and worse prognosis.[5]
Another team discovered that Parkinson's Disease patients with higher abundance of Christensenellaceae had worse nonmotor symptoms. Increased Lactobacillaceae and decreased Lachnospiraceae also correlated with worse cases of intellectual impairment, nonmotor symptoms, gait instability, and posture problems.[4]
Interactions with L-Dopa
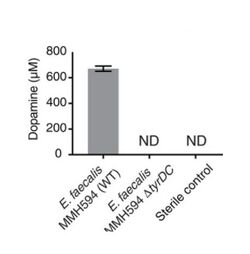
Levodopa (L-dopa) is widely used in the treatment of Parkinson's Disease. Despite its efficacy, up to 56% of administered L-dopa fails to reach the brain because it is metabolized in other pathways. Gut bacteria are suspected to play a large role in loss of L-dopa function; as such, identifying any microbes responsible for metabolizing L-dopa is crucial.[6]
Genome mining was used to identify a protein, tyrosine decarboxylase (TyrDC), in human gut Enterococcus faecalis capable of decarboxylating L-dopa and producing dopamine. TyrDC's role was confirmed by experiments that found knockout of the TyrDC gene drastically reduced Enterococcus faecalis dopamine production. Furthermore, enzyme assays with TyrDC using both L-dopa and the preferred substrate tyrosine showed that TyrDC was capable of decarboxylating both substrates simultaneously, indicating that substrate competition would not hinder TyrDC's affinity for L-dopa.[6]
References
- ↑ 1.0 1.1 Keshavarzian,Ali, Stefan J. Green, Phillip A. Engen, Robin M. Voigt, Ankur Naqib, Christopher B. Forsyth, Ece Mutlu, Kathleen M. Shannon. (2015). "Colonic bacterial composition in Parkinson's disease." Movement Disorders, vol. 30(10), 1351-1360. https://doi.org/10.1002/mds.26307
- ↑ Sampson, Timothy. (2020). "The Impact of Indigenous Microbes on Parkinson's Disease." Neurobiology of Disease, vol. 135. https://doi.org/10.1016/j.nbd.2019.03.014
- ↑ Quigley, Eamonn. (2017). "Gut microbiome as a clinical tool in gastrointestinal disease management: are we there yet?" Nature Reviews Gastroenterology & Hepatology, vol. 14, 315-320. https://doi.org/10.1038/nrgastro.2017.29
- ↑ 4.0 4.1 Barichella, M., Severgnini, M., Cilia, R., Cassani, E., Bolliri, C., Caronni, S., Ferri, V., Cancello, R., Ceccarani, C., Faierman, S., Pinelli, G., De Bellis, G., Zecca, L., Cereda, E., Consolandi, C. and Pezzoli, G. (2019). "Unraveling Gut Microbiota in Parkinson's Disease and Atypical Parkinsonism. Movement Disorders, vol. 34, 396-405. https://doi.org/10.1002/mds.27581
- ↑ 5.0 5.1 Scheperjans, Filip, Velma Aho, Pedro A. B. Pereira, Kaisa Koskinen, Lars Paulin, Eero Pekkonen, Elena Haapaniemi, Seppo Kaakkola, Johanna Eerola-Rautio, Marjatta Pohja, Esko Kinnunen, Kari Murros, Petri Auvinen. (2014). "Gut microbiota are related to Parkinson's disease and clinical phenotype." Movement Disorders, vol. 30(3), 350-358. https://doi.org/10.1002/mds.26069
- ↑ 6.0 6.1 Rekdal, Vayu, Elizabeth Bess, Jordan Bisanz, Peter Turnbaugh, & Emily Balskus. (2019). "Discovery and inhibition of an interspecies gut bacterial pathway for Levodopa metabolism." Science, vol. 364(6445). 10.1126/science.aau6323
Authored for BIOL 238 Microbiology, taught by Joan Slonczewski, 2022, Kenyon College